Objectives
In this lab, you will explore a tool called Kallisto which quantifies abundances of transcripts by pseudomapping RNA-Seq data. Simulated RNA-seq data will be provided to you; the data contains 75 bp paired-end reads that have been generated in silico to replicate real gene count data from Drosophila. The data simulates two biological groups with three biological replicates per group (6 samples total). The objectives of this lab is mainly to:
Learn how Kallisto works and how to use it.
Introduction
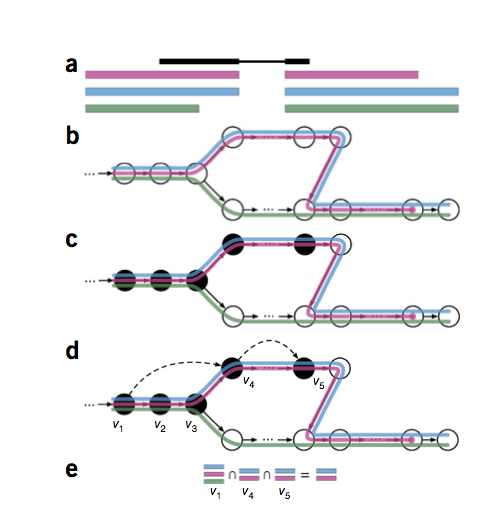
Kallisto is a super fast tool for transcript quantification. It gets it speed by skipping the alignment step. Instead of actually aligning reads to a transcriptome, it identifies transcripts that a read is compatible with in order to quantify the transcript. This process is called pseudoalignment/pseudomapping.
Typically, after quality assessment, the first two steps of RNA-Seq workflow are mapping, followed by quantification of genes/transcripts. Mapping with a mapper can take a 6+ hours for a very large file. Quantification can then take several more hours. Kallisto avoids the mapping step and through a process called pseudoalignment/ pseudomapping, it proceeds directly to the quantification step. It is reported that Kallisto can quantify 30 million human reads in less than 3 minutes on a mac laptop.
Get your data
Six raw data files have been provided for all our further RNA-seq analysis:
- c1_r1, c1_r2, c1_r3 from the first biological condition
- c2_r1, c2_r2, and c2_r3 from the second biological condition
Get set up for the exercises
cds cd my_rnaseq_course cd day_2/kallisto_exercise
module spider kallisto module load intel/17.0.4 module load hdf5/1.8.16 module load kallisto/0.43.1
Part 1. Create a index of your reference
NO NEED TO RUN THIS NOW- YOUR INDEX HAS ALREADY BEEN BUILT!
kallisto index -i transcripts.idx transcripts.fasta
Submit to the TACC queue or run in an idev shell
Create a commands file and use launcher_creator.py followed by sbatch.
nano commands.quant kallisto quant -i ../reference/transcripts.idx -b 100 -o GSM794483_kallisto ../data/GSM794483_C1_R1_1.fq ../data/GSM794483_C1_R1_2.fq kallisto quant -i ../reference/transcripts.idx -b 100 -o GSM794484_kallisto ../data/GSM794484_C1_R2_1.fq ../data/GSM794484_C1_R2_2.fq kallisto quant -i ../reference/transcripts.idx -b 100 -o GSM794485_kallisto ../data/GSM794485_C1_R3_1.fq ../data/GSM794485_C1_R3_2.fq kallisto quant -i ../reference/transcripts.idx -b 100 -o GSM794486_kallisto ../data/GSM794486_C2_R1_1.fq ../data/GSM794486_C2_R1_2.fq kallisto quant -i ../reference/transcripts.idx -b 100 -o GSM794487_kallisto ../data/GSM794487_C2_R2_1.fq ../data/GSM794487_C2_R2_2.fq kallisto quant -i ../reference/transcripts.idx -b 100 -o GSM794488_kallisto ../data/GSM794488_C2_R3_1.fq ../data/GSM794488_C2_R3_2.fq
Output files:
abundances.tsv: A tsv file containing raw read count and a normalized expression value for each transcript.
Back to Course Outline