...
This should be second nature by now
...
Stage the alignment data
First stage the sample datasets and references we will use.
...
| Tip | |||||
|---|---|---|---|---|---|
The BioITeam maintains a set of reference indexes for many common organisms and aligners. They can be found in aligner-specific sub-directories of the /work/projects/BioITeam/ref_genome area. E.g.:
|
Exercise #1: BWA global alignment – Yeast ChIP-seq
...
| Expand | |||||
|---|---|---|---|---|---|
| |||||
The last few lines of bwa's execution output should look something like this:
So the R2 alignment took only 21 seconds, or about 10 times as fast as with only 1 one processing thread. |
Next we use the bwa sampe command to pair the reads and output SAM format data. Just type that command in with no arguments to see its usage.
For this command you provide the same reference index prefix as for bwa aln, along with the two .sai files and the two original FASTQ files. Also, bwa writes its output to standard output, so redirect that to a .sam file.
...
| Code Block | ||||
|---|---|---|---|---|
| ||||
bwa sampe sacCer3/sacCer3.fa yeast_R1.sai yeast_R2.sai fastq/Sample_Yeast_L005_R1.cat.fastq.gz fastq/Sample_Yeast_L005_R2.cat.fastq.gz > yeast_pairedend.sam |
You did it! You should now have a SAM file (yeast_pairedend.sam) that contains the alignments. It's just a text file, so take a look with head, more, less, tail, or whatever you feel like. In the next section, with samtools, you'll learn some additional ways to analyze the data once you create a BAM file.
Exercise: What kind of information is in the first lines of the SAM file?
| Expand | ||
|---|---|---|
| ||
| The SAM or BAM file has a number of header lines, which all start with an at sign ( @ ). The @SQ lines describe each contig and its length. There is also a @PG line that describes the way the bwa sampe was performed. |
...
| Expand | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| ||||||||||
This looks for the pattern '^HWI' which is the start of every read name (which starts every alignment record).
Or use the -v (invert) option to tell grep to print all lines that don't match a particular pattern; here, here all the header lines starting that start with @.
|
...
Exercise: How many sequences were in the R1 and R2 FASTQ files combined?
| Expand | ||
|---|---|---|
| ||
|
| Expand | ||
|---|---|---|
| ||
| There were a total of 1,184,360 original sequences |
...
- Do both R1 and R2 reads have separate alignment records?
- Does the SAM file contain both aligned mapped and un-aligned mapped reads?
- What is the order of the alignment records in this SAM file?
| Expand | ||
|---|---|---|
| ||
| Do both Both R1 and R2 reads must have separate alignment records ?yes, they must,because there were 1,184,360 R1+R2 reads and an equalthe same number of alignment records. Does theThe SAM file must contain both alignedmapped and un- aligned reads?yes, it mustmapped reads, because there were 1,184,360 R1+R2 reads and an equaland the same number of alignment records What is the order of the alignment records in this SAM file?. Alignment records the namesoccur in the exactsame read-name order as they did in the FASTQ, except that they come in pairs the. The R1 read comes first1st, then itsthe corresponding R2 this ordering. This is called read name ordering. |
Using cut to isolate fields
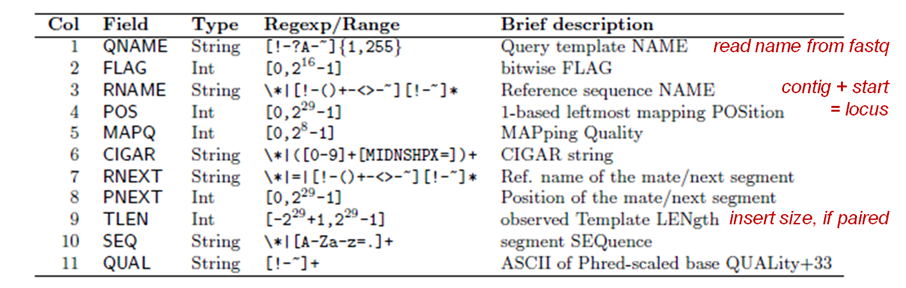
Recall the format of a SAM/BAM file alignment record:
Suppose you wanted to look only at field 3 (contig name) values in the SAM file. You can do this with the handy cut command. Below is a simple example where you're asking cut to display the 3rd column value for the last 10 alignment records.
...
By default cut assumes the field delimiter is Tab, which is the delimiter used in the majority of NGS file formats. You can , of course, specify a different delimiter with the -d option.
You can also specify a range of fields, and mix adjacent and non-adjacent fields. This displays fields 2 through 6, field 9, and all fields starting with 12 the 12th (SAM tag fields).
| Code Block | ||||
|---|---|---|---|---|
| ||||
tail yeast_pairedend.sam | cut -f 2-6,9,12- |
You may have noticed that some alignment records contain contig names (e.g. chrV) in field 3 while others contain an asterisk ( * ). Usually the * means the record didn't alignmap. (This isn't always true – later you'll see how to properly distinguish between mapped and unmapped reads using samtools.) We're going to use this heuristic along with cut to see about how many records represent aligned sequences.
First we need to make sure that we don't look at fields in the SAM header lines. We're going to end up with a series of pipe operations, and the best way to make sure you're on track is to enter them one at a time piping to head:
| Code Block | ||||
|---|---|---|---|---|
| ||||
# the ^HWI pattern matches lines starting with HWI (the start of all read names in column 1) grep -P '^HWI' yeast_pairedend.sam | head |
...
| Expand | ||
|---|---|---|
| ||
| Recall that these are 100 bp reads and we did not remove adapter contamination. There will be a distribution of fragment sizes – some will be short – and those short fragments may not align without adapter removal (e.g. with fastx_trimmer or cutadapt). |
Exercise
...
#2: Simple SAMtools Utilities
We have used several alignment methods that all generate results in the form of the near-universal SAM/BAM file format. The SAMtools program is a ubiquitously used set of tools that allow a user to manipulate SAM/BAM files in many different ways, ranging from simple tasks (like SAM/BAM interconversion) to more complex functions (like removal of PCR duplicates). It is available in the TACC module system in the typical fashion.
...