...
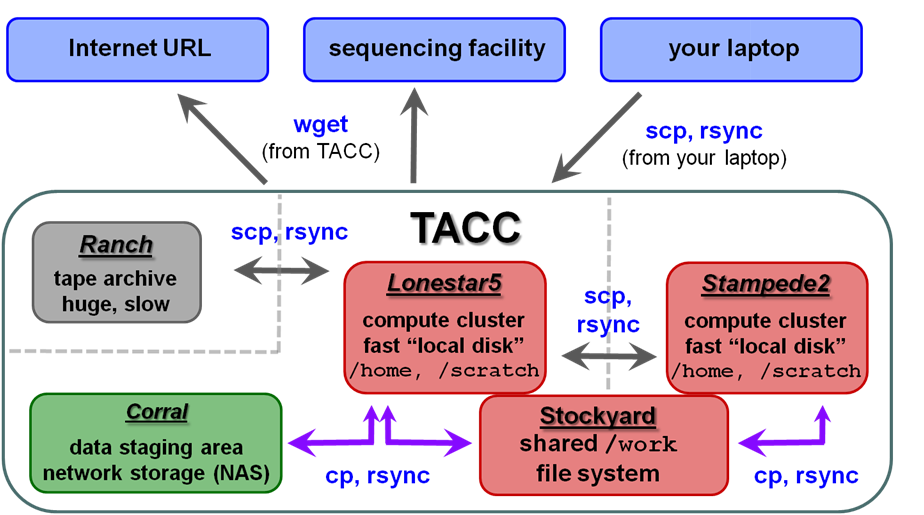
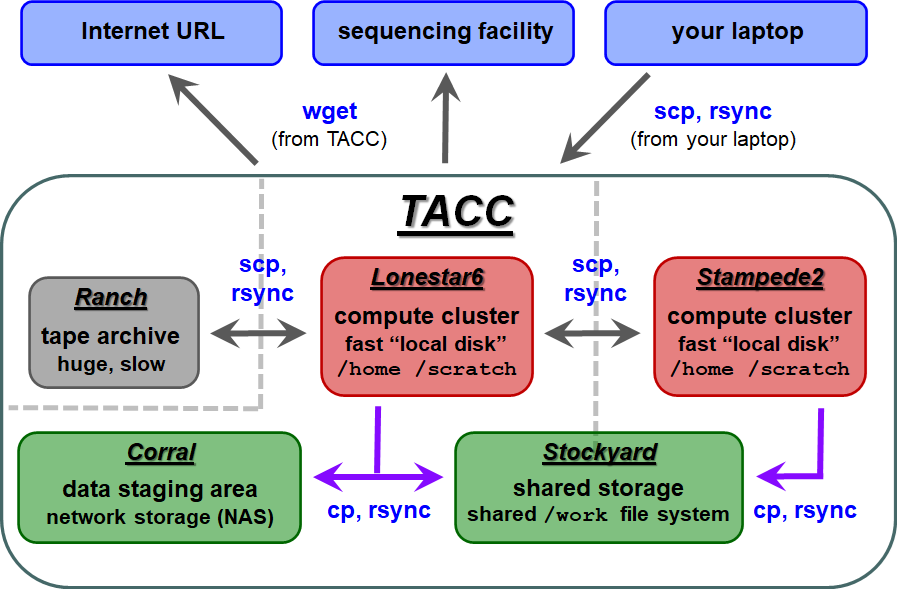
| TACC storage areas and Linux commands to access data (all commands to be executed at TACC except laptop-to-TACC copies, which must be executed on your laptop) |
Local file systems
There are 3 local file systems available on any TACC compute cluster (Lonestar5stampede2, stampede2 lonestar6, etc.), each with different characteristics. All these local file systems are very fast and set up for parallel I/O (Lustre file system).
On ls5 lonestar6 these local file systems have the following characteristics:
| Home | Work | Scratch | |
|---|---|---|---|
| quota | 10 GB | 1024 GB = 1 TB | 2+ PB (basically infinite) |
| policy | backed up | not backed up, not purged | not backed up, purged if not accessed recently (~10 days) |
| access command | cd | cdw | cds |
| environment variable | $HOME | $WORK (different sub-directory for each cluster) $STOCKYARD (root of the shared Work file system )$WORK (different sub-directory for each cluster ) | $SCRATCH |
| root file system | /home | /work | /scratch |
| use for | Small files such as scripts that you don't want to lose. | Medium-sized artifacts files you don't want to copy over all the time. For example, custom programs you install (these can get large), or annotation file used for analysis. | Large files accessed from batch jobs. Your starting files will be copied here from somewhere else, and your final results files will be copied elsewhere (e.g. stockyard, corral, or your BRCF POD), or your organization's storage area. |
When you first login, the system gives you information about disk quota quotas and your compute allocation quota:balance in "SU" (system units).
| Code Block |
|---|
--------------------- Project balances for user abattenh ---------------------- | Name Avail SUs Expires | Name Avail SUs Expires | | genomeAnalysisOTH21095 673 2021-03-31 | BioinformaticsResour 1001000 20202022-0609-30 | | UT-2015-05-18MCB21106 1000 2021-03-31 | DNAdenovo 995 2022-12-31 | | OTH21164 4969 2021-03-31 | | CancerGenetics 48561000 20202022-0912-3031 | A-cm10OTH21037 4224 8867 20202022-12-31 | ------------------------ Disk quotas for user abattenh ------------------------ | Disk Usage (GB) Limit %Used File Usage Limit %Used | | /home1 0.0 1011.07 0.1000 52 153 10000000 0.0200 | | /work 614169.50 1024.0 6016.0150 6109479362 3000000 2.04 | | /scratch 2676.6 0.0 0.00 32442 0 0.00 | -65 | --------------------------------------------------------------------------------------------------- |
changing TACC file systems
When you first login, you start in your home Home directory. Use these the cd, cdw and cds commands to change to your other file systems. Notice how your command prompt helpfully changes to show your location.
...
| Tip |
|---|
The cd (change directory) command with no arguments takes you to your home directory on any Linux/Unix system. The cdw and cds commands are specific to the TACC environment. |
...
TACC compute clusters now share a common Work file system called stockyard. So files in your Work area do not have to be copied, for example from ls5 to stampede2 to ls6 – they can be accessed directly from either cluster.
...
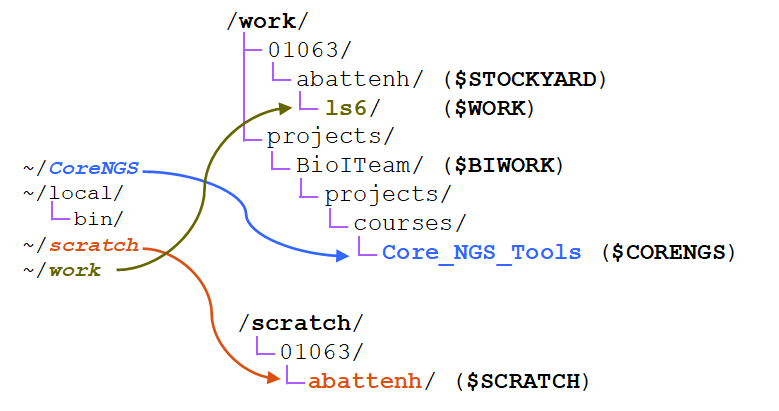
- $STOCKYARD - This refers to the root of your shared Work area
- e.g. /work/01063/abattenh
- e.g. /work/01063/abattenh
- $WORK - Refers to a sub-directory of the shared Work area that is different for different clusters, e.g.:
- /work/01063/abattenh/lonestarls6 on ls5 lonestar6
- /work/01063/abattenh/stampede2 on stampede2
...
corral is a gigantic (multiple PB) storage system (spinning disk) where researchers can store data. UT researchers may request up to 5 TB of corral storage through the normal TACC allocation request process. Additional space on corral can be rented for ~$85~$80/TB/year.
The UT/Austin BioInformatics Team also has an older, common directory area on corral.
...
...
ls /corral-repl/utexas/BioITeamA couple of things to keep in mind regarding corral:
...
ranch is a gigantic (multiple PB) tape archive system where researchers can archive data. All TACC users have an automatice automatic 2 TB ranch allocation. UT researchers may request larger (multi-TB) ranch storage allocations through the normal TACC allocation request process.
...
Once that data is staged to the ranch disk it can be copied to other places. However, the ranch file system is not mounted as a local file system from the stampede2 or ls5 ls6 clusters. So remote copy commands are always needed to copy data to and from ranch (e.g. scp, rsync).
Staging your data
So, your sequencing center has some data for you. They may send you a list of web or FTP links to use to download the data.
The first task is to get this sequencing data to a permanent storage area. This should not be your laptop! corral (or stockyard) is a great place for it, or a server maintained by your lab or company.
Here's an example of a "best practice". Wherever your permanent storage area is, it should have a rational sub-directory structure that reflects its contents. It's easy to process a few NGS datasets, but when they start multiplying like tribbles, good organization and naming conventions will be the only thing standing between you and utter chaos!
For example:
- original – for original sequencing data (compressed FASTQ files)
- sub-directories named, for example, by year_month.<project_name>
- aligned – for alignment artifacts (BAM files, etc)
- sub-directories named, e.g., by year_month.<project_name>
- analysis – further downstream analysis
- reasonably named sub-directories, often by project
- genome – reference genomes and other annotation files used in alignment and analysis
- sub-directories for different reference genomes
- e.g. ucsc/hg19, ucsc/sacCer3, mirbase/v20
- code – for scripts and programs you and others in your organization write
- ideally maintained in a version control system such as git, subversion or cvs.
- easiest to name sub-directories for people.
Download from a link – wget
Well, you don't have a desktop at TACC to "Save as" to, so what to do with a link? The wget program knows how to access web URLs such as http, https and ftp.
...
wget
Get ready to run wget from the directory where you want to put the data.
Don't press Enter after the wget command – just put a space after it.
| Code Block | ||||
|---|---|---|---|---|
| ||||
mkdir -p $WORK/archive/original/2020.core_ngs
cd $WORK/archive/original/2020.core_ngs
wget |
Here are two web links:
...
About file systems
File systems are storage areas where files and directories are arranged in a hierarchy. Any computer can have one or more file systems "mounted" (accessible as local storage). The df command can be used on any Unix system to show all the "top-level" mounted file systems. TACC has a lot of temporary file systems, so lets just look at the first 15 and tell df to use "human readable" size formatting with the -h option:
| Code Block | ||
|---|---|---|
| ||
df -h | head -15 |
The rightmost "Mounted on" column give the top-level access path. Find /home1, /work, and /scratch and note their sizes!
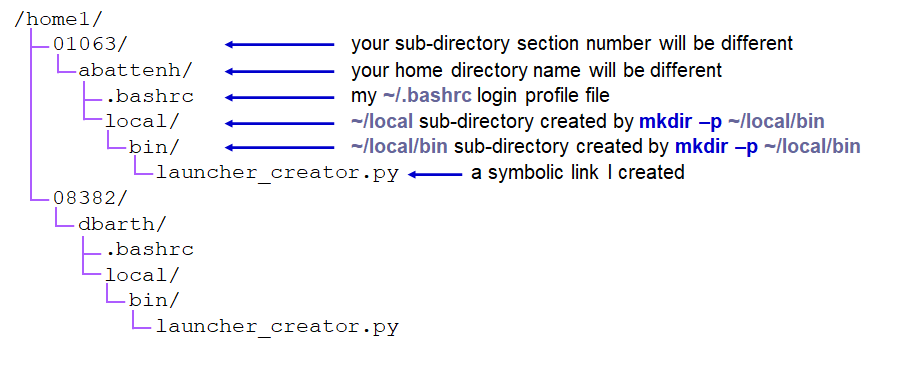
What do we mean by "hierarchy"? It is like a tree, with the root file system as the trunk, sub-directories as branches, sub-sub-directories as branches from branches (and so forth), with files as leaves off any branch.
your TACC Home directory |
But everyone has a Home directory, so you must only be seeing a part of the Home directory hierarchy. To see the absolute path of a directory you're in, use the pwd -P command. Note that absolute paths always start with a forward slash ( / ) denoting its root file system.
| Code Block | ||
|---|---|---|
| ||
pwd -P
# will show something like this
# /home1/01063/abattenh |
That shows you that your Home directory (~) is actually 3 levels down in the /home1 hierarchy:
| part of the TACC Home file system using absolute paths |
Here's a depiction of the three file systems as seen from your Home directory ( ~ ), showing where the path-valued environment variables represent, and where the three symbolic links (~/CoreNGS, ~/scratch, ~/work) you created in your Home directory point. Notice that both the Work and Scratch file systems have a top-level hierarchy like we saw in Home above.
Staging your data
So, your sequencing center has some data for you. They may send you a list of web or FTP links to use to download the data.
The first task is to get this sequencing data to a permanent storage area. This should not be your laptop! corral (or stockyard) is a great place for it, or a server maintained by your lab or company.
Here's an example of a "best practice". Wherever your permanent storage area is, it should have a rational sub-directory structure that reflects its contents. It's easy to process a few NGS datasets, but when they start multiplying like tribbles, good organization and naming conventions will be the only thing standing between you and utter chaos!
For example:
- original – for original sequencing data (compressed FASTQ files)
- sub-directories named, for example, by year_month.<sequencing run/job or project name>
- aligned – for alignment data (BAM files, etc)
- sub-directories named, e.g., by year_month.<project_name>
- analysis – further downstream analysis
- reasonably named sub-directories, often by project
- refs – reference genomes and other annotation files used in alignment and analysis
- sub-directories for different reference genomes and aligners
- e.g. ucsc/hg38/star, ucsc/sacCer3/bwa, mirbase/v20/bowtie2
- code – for scripts and programs you and others in your organization write
- ideally maintained in a version control system such as git, subversion or cvs.
- can have separate sub-directories for people, or various shared repositories.
Download from a link – wget
Well, you don't have a desktop at TACC to "Save as" to, so what to do with a link? The wget program knows how to access web URLs such as http, https and ftp.
| Anchor | ||||
|---|---|---|---|---|
|
wget
Get ready to run wget from the directory where you want to put the data.
Don't press Enter after the wget command – just put a space after it.
| Code Block | ||||
|---|---|---|---|---|
| ||||
mkdir -p $SCRATCH/archive/original/2021.core_ngs
cd $SCRATCH/archive/original/2021.core_ngs
wget |
Here are two web links:
- https://web.corral.tacc.utexas.edu/BioinformaticsResource/CoreNGS/yeast_stuff/Sample_Yeast_L005_R1.cat.fastq.gz
- https://web.corral.tacc.utexas.edu/BioinformaticsResource/CoreNGS/yeast_stuff/Sample_Yeast_L005_R2.cat.fastq.gz
Right-click (Windows) or Control+click (Mac) on the 1st link in your browser, then select "Copy link location" from the menu. Now go back to your Terminal. Put your cursor after the space following the wget command then either right-click (Windows), or Paste (Command-V on Mac, Control-V on Windows). The command line to be executed should now look like this:
| Code Block | ||||
|---|---|---|---|---|
| ||||
wget http://web.corral.tacc.utexas.edu/ |
...
BioinformaticsResource/CoreNGS/yeast_stuff/Sample_Yeast_L005_ |
...
R1.cat.fastq.gz |
Right-click (Windows) or Control+click (Mac) on the 1st link in your browser, then select "Copy link location" from the menu. Now go back to your Terminal. Put your cursor after the space following the wget command then either right-click (Windows), or Paste (Command-V on Mac, Control-V on Windows). The command line to be executed should now look like this:
Now press Enter to get the command going. Repeat for the 2nd link. Check that you now see the two files (ls).
| Tip | ||||
|---|---|---|---|---|
| Code Block | ||||
| ||||
| wget http://web.corral.tacc.utexas.edu/BioITeam/yeast_stuff/ By default wget creates a file in the current directory matching the last component of the URL (e.g. Sample_Yeast_L005_R1.cat.fastq.gz |
...
here). You can change the copied file name with wget's -O option. Also note that if you execute the same wget more than once, subsequent local files will be named with a .1, .2, etc. suffix. |
Copy from a corral location - cp or rsync
...
The cp command copies one or more files from a local source to a local destination. It has many options, but the most common form is:
cp [options] <source_file_1> <source_file_2> ... <destination_directory>/
Make a directory in your scratch Scratcharea and copy a single file to it. The trailing slash ( / ) on the destination says it the destination is a directory.
| Code Block | ||||
|---|---|---|---|---|
| ||||
mkdir -p $SCRATCH/data/test1
cp $CORENGS/misc/small.fq $SCRATCH/data/test1/
ls $SCRATCH/data/test1
# or..
mkdir -p ~/scratch/data/test1 # use the symbolic link in your Home directory
cd ~/scratch/data/test1
cp $CORENGS/misc/small.fq .
ls |
Copy an entire directory to your scratch Scratch area. The -r argument option says "recursive".
| Code Block | ||||
|---|---|---|---|---|
| ||||
mkdir -p $SCRATCH/data
cds
cd data
cp -r $CORENGS/general/ general/ |
Exercise: What files were copied over?
| Expand | ||
|---|---|---|
| ||
ls ls general |
| Expand | ||
|---|---|---|
| ||
BEDTools-User-Manual.v4.pdf SAM1.pdf SAM1.v1.4.pdf |
...
rsync is a very complicated program, with many options (http://rsync.samba.org/ftp/rsync/rsync.html). However, if you use the recipe shown here for directories, it's hard to go wrong:
rsync -avW local/path/to/source_directory/ local/path/to/destination_directory/
Both the source and target directories are local (in some file system accessible directly from ls5 lonestar6). Either full or relative path syntax can be used for both. The -avP avW options above stand for:
- -a means "archive mode", which implies the following options (and a few others)
- -p – preserve file permissions
- -t – preserve file times
- -l – copy symbolic links as links
- -r – recursively copy sub-directories
- -v means verbose
- -W means transfer Whole file only
- Normally the rsync algorithm compares the contents of files that need to be copied and only transfers the different parts.
- For large files and binary files, figuring out what has changed (diff-ing) can take more time than just copying the whole file.
- The -W option disables file content comparisons (skips diff-ing).
...
| Tip | ||
|---|---|---|
| ||
The trailing slash ( / ) on the source and destination directories are very important for rsync (and for other Linux copy commands also)! rsync will create the last directory level for you, but earlier levels must already exist. |
...
| Code Block | ||||
|---|---|---|---|---|
| ||||
mkdir -p $SCRATCH/data cds rsync -avWPavrW $CORENGS/custom_tracks/ data/custom_tracks/ |
...
| Expand | ||
|---|---|---|
| ||
ls $SCRATCH/data/custom_tracks |
Now repeat the rsync and see the difference.
...
| Code Block | ||
|---|---|---|
| ||
rsync -avWPavrW /work/projects/BioITeam/projects/courses/Core_NGS_Tools/custom_tracks/ data/custom_tracks/ |
...
| Tip |
|---|
The bash shell has several convenient line editing features:
|
Copy from a remote computer - scp or rsync
...
| Code Block | ||
|---|---|---|
| ||
mkdir -p $SCRATCH/data/test1 cds cat $CORENGS/tacc/dragonfly_access.txt cds mkdir -p data/test2 scp corengstools@dragonfly.icmb.utexas.edu:~/custom_tracks/progeria_ctcf.vcf.gz ./data/test1test2/ lstree .$SCRATCH/data/test1test2 |
Notes:
- The 1st time you access a new host the SSH security prompt will appear
- You will be prompted for your remote host password
- The -r recursive argument works for scp also, just like for cp
...
- The tilde ( ~ ) at the start of the path means "relative to my home directory"
- We use the tilde ( ~ ) in the destination to traverse the ~/scratch symbolic link in your home directory.
| Code Block | |||||
|---|---|---|---|---|---|
| |||||
cat $CORENGS/tacc/dragonfly_access.txt rsync -avWPavrW corengstools@dragonfly.icmb.utexas.edu:~/custom_tracks/ ~/scratch/data/custom_tracks/ |
...
| Expand | ||
|---|---|---|
| ||
No, because all the source files were already present in the destination directory (you copied the same files earlier) with the same names, file sizes and timestamps. So rsync had nothing to do! |
...
Here's a fun scavenger hunt for more practice . Issue of the following commands to get practice what you've learned so far:.
| Expand | ||
|---|---|---|
| ||
Hit Tab Tab as much as possible to save typing! |
To get started:started:
| Code Block | ||
|---|---|---|
| ||
cd
cp -r /work/projects/BioITeam/projects/courses/Core_NGS_Tools/linuxpractice/what what
# or using the $CORENGS environment variable | ||
| Code Block | ||
| ||
cd
cp -r $CORENGS/linuxpractice/what what
cd what
cat readme |
Where are you when you're all done?
| Expand | ||
|---|---|---|
| ||
|
...
| Expand | |||||
|---|---|---|---|---|---|
| |||||
From inside your ~/what/starts directory:
|
| Expand | |||||
|---|---|---|---|---|---|
| |||||
From inside your ~/what/starts/here directory:
|
...
| Expand | |||||
|---|---|---|---|---|---|
| |||||
From inside your ~/what/starts/here/changes directory:
The path to the directory you're in now should be: ~/what/starts/here/changes/the/world |