Overview
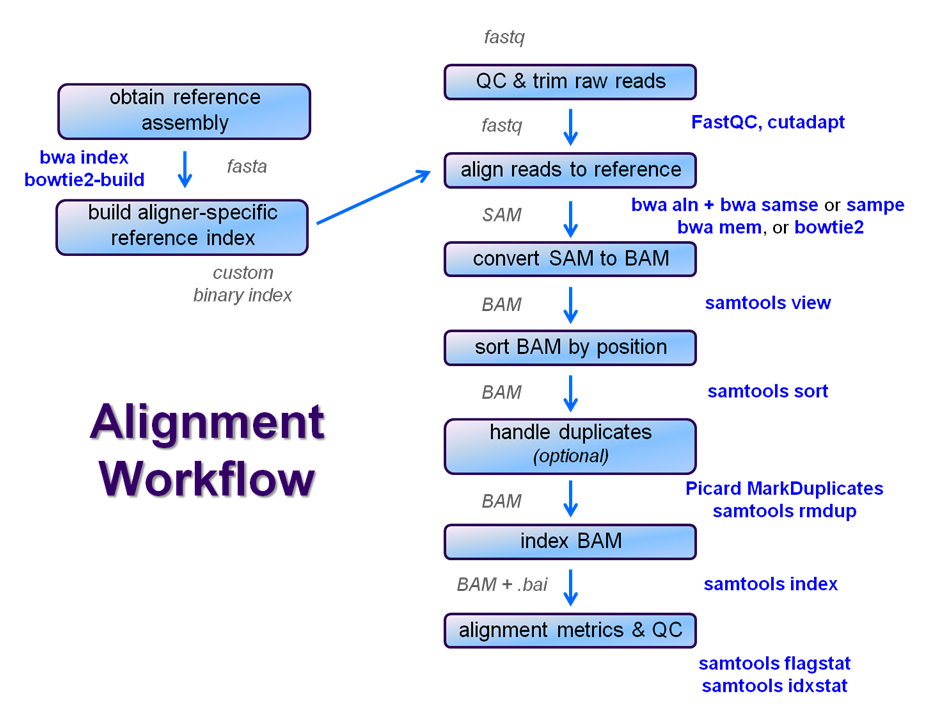
After raw sequence files are generated (in FASTQ format), quality-checked, and pre-processed in some way, the next step in most many NGS pipelines is mapping to a reference genome.
For individual sequences , it is common to use a tool like BLAST to identify genes or species of origin. However , a normal NGS dataset will have tens to hundreds of millions of sequences, which BLAST and similar tools are not designed to handle. Thus , a large set of computational tools have been developed to quickly , align each read to its best location , (if any, ) in a reference.
Even though many mapping tools exist, a few individual programs have a dominant "market share" of the NGS world. In this section, we will primarily focus on two of the most versatile mappersgeneral-purpose ones: BWA and Bowtie2, (the latter being part of the Tuxedo suite which includes the transcriptome-aware RNA-seq aligner Tophat2 as well as other downstream quantifiaction tools).
...
Stage the alignment data
First connect to stampede2.tacc.utexas.edu
...
and start an idev session. This should be second nature by now 
Stage the alignment data
| Code Block |
|---|
| language | bash |
|---|
| title | Start an idev session |
|---|
|
idev -p normal -m 180 -A UT-2015-05-18 -N 1 -n 68 |
Then First stage the sample datasets and references we will use.
| Code Block |
|---|
| language | bash |
|---|
| title | Get the alignment exercises files |
|---|
|
mkdir -p $SCRATCH/core_ngs/alignmentreferences/fastqfasta
mkdir -p $SCRATCH/core_ngs/referencesalignment/fastafastq
cp $CORENGS/alignmentreferences/*fastq.gzfa $SCRATCH/core_ngs/alignmentreferences/fastqfasta/
cp $CORENGS/referencesalignment/*fastq.*gz $SCRATCH/core_ngs/references/fasta//alignment/fastq/
cd $SCRATCH/core_ngs/alignment/fastq |
These are descriptions of the FASTQ files we copied:
| File Name | Description | Sample |
|---|
| Sample_Yeast_L005_R1.cat.fastq.gz | Paired-end Illumina, First of pair, FASTQ | Yeast ChIP-seq |
| Sample_Yeast_L005_R2.cat.fastq.gz | Paired-end Illumina, Second of pair, FASTQ | Yeast ChIP-seq |
| human_rnaseq.fastq.gz | Paired-end Illumina, First of pair only, FASTQ | Human RNA-seq |
| human_mirnaseq.fastq.gz | Single-end Illumina, FASTQ | Human microRNA-seq |
| cholera_rnaseq.fastq.gz | Single-end Illumina, FASTQ | V. cholerae RNA-seq |
Reference Genomes
Before we get to alignment, we need a reference to align to. This is usually an organism's genome, but can also be any set of names sequences, such as a transcriptome or other set of genes.
...
Searching genomes is computationally hard work and takes a long time if done on un-indexed, linear genomic sequence. So aligners require that references first be indexed to accelerate lookup. The aligners we are using each require a different index, but use the same method (the Burrows-Wheeler Transform) to get the job done.
Building a reference index involves taking a FASTA file as input, with each chromosome (or contig contig (contiguous string of bases, e.g. a chromosome) as a separate FASTA entry, and producing an aligner-specific set of files as output. Those output index files are then used to perform the sequence alignment, and alignments are reported using coordinates referencing names and offset positions based on the original FASTA file contig entries.
We can quickly index the references for the yeast genome, the human miRNAs, and the V. cholerae genome, because they are all small, so we'll grab build each index from the appropriate FASTA files for yeast and human miRNAs references and build each index right before we use them. We will also obtain the special GenBank file that contains both the V. cholerae genome sequence and annotations (a .gbk file). These FASTA files, which you staged above, are
hg19 is way too big for us to index here so we will use an existing set of BWA hg19 index files located at:
| Code Block |
|---|
| language | bash |
|---|
| title | Reference FASTA locations |
|---|
|
/work/projects/BioITeam/projects/courses/Core_NGS_Tools/references/sacCer3.fa
/work/projects/BioITeam/projects/courses/Core_NGS_Tools/references/hairpin_cDNA_hsa.fa
/work/projects/BioITeam/projects/courses/Core_NGS_Tools/references/vibCho.O395.gbk |
hg19 is way too big for us to index here so we will use an existing set of BWA hg19 index files located at:
|
/work2 |
| Code Block |
|---|
| language | bash |
|---|
| title | BWA hg19 index location |
|---|
|
/work/projects/BioITeam/ref_genome/bwa/bwtsw/hg19 |
| Tip |
|---|
The BioITeam maintains a set of reference indexes for many common organisms and aligners. They can be found in aligner-specific sub-directories of the /workwork2/projects/BioITeam/ref_genome area. E.g.: | Code Block |
|---|
| /workwork2/projects/BioITeam/ref_genome/
bowtie2/
bwa/
hisat2/
kallisto/
star/
tophat/ |
|
Exercise #1: BWA global alignment – Yeast ChIP-seq
Overview ChIP-seq alignment workflow with BWA
We will perform a global alignment of the paired-end Yeast ChIP-seq sequences using bwa. This workflow has the following steps:
- Trim the FASTQ sequences down to 50 with fastx_clipper
- this removes most of any 5' adapter contamination without the fuss of specific adapter trimming w/cutadapt
- Prepare the sacCer3 reference index for bwa using bwa index
- this is done once, and re-used for later alignments
- Perform a global bwa alignment on the R1 reads (bwa aln) producing a BWA-specific binary .sai intermediate file
- Perform a global bwa alignment on the R2 reads (bwa aln) producing a BWA-specific binary .sai intermediate file
- Perform pairing of the separately aligned reads and report the alignments in SAM format using bwa sampe
- Convert the SAM file to a BAM file (samtools view)
- Sort the BAM file by genomic location (samtools sort)
- Index the BAM file (samtools index)
- Gather simple alignment statistics (samtools flagstat and samtools idxstat)
We're going to skip the trimming step for now and see how it goes. We'll perform steps 2 - 5 now and leave samtools for the next exercise since steps 6 - 10 are common to nearly all post-alignment workflows.
Introducing BWA
Like other tools you've worked with so far, you first need to load bwa using the module system. Do that now, and then enter bwa with no arguments to view the top-level help page (many NGS tools will provide some help when called with no arguments).
| Code Block |
|---|
|
module load bwa
bwa |
| Expand |
|---|
|
| Code Block |
|---|
Program: bwa (alignment via Burrows-Wheeler transformation)
Version: 0.7.12-r1039
Contact: Heng Li <lh3@sanger.ac.uk>
Usage: bwa <command> [options]
Command: index index sequences in the FASTA format
mem BWA-MEM algorithm
fastmap identify super-maximal exact matches
pemerge merge overlapping paired ends (EXPERIMENTAL)
aln gapped/ungapped alignment
samse generate alignment (single ended)
sampe generate alignment (paired ended)
bwasw BWA-SW for long queries
shm manage indices in shared memory
fa2pac convert FASTA to PAC format
pac2bwt generate BWT from PAC
pac2bwtgen alternative algorithm for generating BWT
bwtupdate update .bwt to the new format
bwt2sa generate SA from BWT and Occ
Note: To use BWA, you need to first index the genome with `bwa index'.
There are three alignment algorithms in BWA: `mem', `bwasw', and
`aln/samse/sampe'. If you are not sure which to use, try `bwa mem'
first. Please `man ./bwa.1' for the manual. |
|
As you can see, bwa include many sub-commands that perform the tasks we are interested in.
Building the BWA sacCer3 index
We will index the genome with the bwa index command. Type bwa index with no arguments to see usage for this sub-command.
| Code Block |
|---|
Usage: bwa index [-a bwtsw|is] [-c] <in.fasta>
Options: -a STR BWT construction algorithm: bwtsw or is [auto]
-p STR prefix of the index [same as fasta name]
-b INT block size for the bwtsw algorithm (effective with -a bwtsw) [10000000]
-6 index files named as <in.fasta>.64.* instead of <in.fasta>.*
Warning: `-a bwtsw' does not work for short genomes, while `-a is' and
`-a div' do not work not for long genomes. Please choose `-a'
according to the length of the genome. |
Based on the usage description, we only need to specify two things:
- the name of the FASTA file
- whether to use the bwtsw or is algorithm for indexing
Since sacCer3 is relative large (~12 Mbp) we will specify bwtsw as the indexing option (as indicated by the "Warning" message), and the name of the FASTA file is sacCer3.fa.
Importantly, the output of this command is a group of files that are all required together as the index. So, within our references directory, we will create another directory called references/bwa/sacCer3 and build the index there. To remind ourselves which FASTA was used to build the index, we create a symbolic link to our references/fasta/sacCer3.fa file (note the use of the ../.. relative path syntax).
| Code Block |
|---|
| language | bash |
|---|
| title | Prepare BWA reference directory for sacCer3 |
|---|
|
mkdir -p $SCRATCH/core_ngs/references/bwa/sacCer3
cd $SCRATCH/core_ngs/references/bwa/sacCer3
ln -s ../../fasta/sacCer3.fa
ls -l |
Now execute the bwa index command.
| Code Block |
|---|
| language | bash |
|---|
| title | Build BWA index for sacCer3 |
|---|
|
bwa index -a bwtsw sacCer3.fa |
Since the yeast genome is not large when compared to human, this should not take long to execute (otherwise we would do it as a batch job). When it is complete you should see a set of index files like this:
| Code Block |
|---|
| title | BWA index files for sacCer3 |
|---|
|
sacCer3.fa
sacCer3.fa.amb
sacCer3.fa.ann
sacCer3.fa.bwt
sacCer3.fa.pac
sacCer3.fa.sa |
Exploring the FASTA with grep
It is often useful to know what chromosomes/contigs are in a FASTA file before you start an alignment so that you're familiar with the contig naming convention – and to verify that it's the one you expect. For example, chromosome 1 is specified differently in different references and organisms: chr1 (USCS human), chrI (UCSC yeast), or just 1 (Ensembl human GRCh37).
A FASTA file consists of a number of contig name entries, each one starting with a right carat ( > ) character, followed by many lines of base characters. E.g.:
| Code Block |
|---|
>chrI
CCACACCACACCCACACACCCACACACCACACCACACACCACACCACACC
CACACACACACATCCTAACACTACCCTAACACAGCCCTAATCTAACCCTG
GCCAACCTGTCTCTCAACTTACCCTCCATTACCCTGCCTCCACTCGTTAC
CCTGTCCCATTCAACCATACCACTCCGAACCACCATCCATCCCTCTACTT |
How do we dig out just the lines that have the contig names and ignore all the sequences? Well, the contig name lines all follow the pattern above, and since the > character is not a valid base, it will never appear on a sequence line.
We've discovered a pattern (also known as a regular expression) to use in searching, and the command line tool that does regular expression matching is grep (general regular expression parser). Read more about grep here: Advanced commands: grep.
Regular expressions are so powerful that nearly every modern computer language includes a "regex" module of some sort. There are many online tutorials for regular expressions, and several slightly different "flavors" of them. But the most common is the Perl style (http://perldoc.perl.org/perlretut.html), which was one of the fist and still the most powerful (there's a reason Perl Perl used extensively when assembling the human genome). We're only going to use the most simple of regular expressions here, but learning more about them will pay handsome dividends for you in the future.
Here's how to execute grep to list contig names in a FASTA file.
| Code Block |
|---|
| language | bash |
|---|
| title | grep to match contig names in a FASTA file |
|---|
|
grep -P '^>' sacCer3.fa | more |
Notes:
- The -P option tells grep to Perl-style regular expression patterns.
- This makes including special characters like Tab ( \t ), Carriage Return ( \r ) or Linefeed ( \n ) much easier that the default POSIX paterns.
- While it is not required here, it generally doesn't hurt to include this option.
'^>' is the regular expression describing the pattern we're looking for (described below)
- sacCer3.fa is the file to search.
- lines with text that match our pattern will be written to standard output
- non matching lines will be omitted
- We pipe to more just in case there are a lot of contig names.
Now down to the nuts and bolts of the pattern: '^>'
First, the single quotes around the pattern – this tells the bash shell to pass the exact string contents to grep.
As part of its friendly command line parsing and evaluation, the shell will often look for special characters on the command line that mean something to it (for example, the $ in front of an environment variable name, like in $SCRATCH). Well, regular expressions treat the $ specially too – but in a completely different way! Those single quotes tell the shell "don't look inside here for special characters – treat this as a literal string and pass it to the program". The shell will obey, will strip the single quotes off the string, and will pass the actual pattern, ^>, to the grep program. (Note that the shell does look inside double quotes ( " ) for certain special signals, such as looking for environment variable names to evaluate. Read more about Quoting in the shell.)
So what does ^> mean to grep? We know that contig name lines always start with a > character, so > is a literal for grep to use in its pattern match.
We might be able to get away with just using this literal alone as our regex, specifying '>' as the command line argument. But for grep, the more specific the pattern, the better. So we constrain where the > can appear on the line. The special carat ( ^ ) character represents "beginning of line". So ^> means "beginning of a line followed by a > character".
Exercise: How many contigs are there in the sacCer3 reference?
...
Exploring FASTA with grep
It is often useful to know what chromosomes/contigs are in a FASTA file before you start an alignment so that you're familiar with the contig naming convention – and to verify that it's the one you expect. For example, chromosome 1 is specified differently in different references and organisms: chr1 (USCS human), chrI (UCSC yeast), or just 1 (Ensembl human GRCh37).
A FASTA file consists of a number of contig name entries, each one starting with a right carat ( > ) character, followed by many lines of base characters. E.g.:
| Code Block |
|---|
>chrI
CCACACCACACCCACACACCCACACACCACACCACACACCACACCACACC
CACACACACACATCCTAACACTACCCTAACACAGCCCTAATCTAACCCTG
GCCAACCTGTCTCTCAACTTACCCTCCATTACCCTGCCTCCACTCGTTAC
CCTGTCCCATTCAACCATACCACTCCGAACCACCATCCATCCCTCTACTT |
How do we dig out just the lines that have the contig names and ignore all the sequences? Well, the contig name lines all follow the pattern above, and since the > character is not a valid base, it will never appear on a sequence line.
We've discovered a pattern (also known as a regular expression) to use in searching, and the command line tool that does regular expression matching is grep (general regular expression parser). Read more about grep here: Advanced commands: grep.
Regular expressions are so powerful that nearly every modern computer language includes a "regex" module of some sort. There are many online tutorials for regular expressions, and several slightly different "flavors" of them. But the most common is the Perl style (http://perldoc.perl.org/perlretut.html), which was one of the fist and still the most powerful (there's a reason Perl was used extensively when assembling the human genome). We're only going to use the most simple of regular expressions here, but learning more about them will pay handsome dividends for you in the future.
Here's how to execute grep to list contig names in a FASTA file.
| Code Block |
|---|
| language | bash |
|---|
| title | grep to match contig names in a FASTA file |
|---|
|
cd $SCRATCH/core_ngs/references/fasta
grep -P '^>' sacCer3.fa | more |
Notes:
- The -P option tells grep to Perl-style regular expression patterns.
- This makes including special characters like Tab ( \t ), Carriage Return ( \r ) or Linefeed ( \n ) much easier that the default POSIX paterns.
- While it is not required here, it generally doesn't hurt to include this option.
'^>' is the regular expression describing the pattern we're looking for (described below)
- sacCer3.fa is the file to search.
- lines with text that match our pattern will be written to standard output
- non matching lines will be omitted
- We pipe to more just in case there are a lot of contig names.
Now down to the nuts and bolts of the pattern: '^>'
First, the single quotes around the pattern – this tells the bash shell to pass the exact string contents to grep.
As part of its friendly command line parsing and evaluation, the shell will often look for special characters on the command line that mean something to it (for example, the $ in front of an environment variable name, like in $SCRATCH). Well, regular expressions treat the $ specially too – but in a completely different way! Those single quotes tell the shell "don't look inside here for special characters – treat this as a literal string and pass it to the program". The shell will obey, will strip the single quotes off the string, and will pass the actual pattern, ^>, to the grep program. (Note that the shell does look inside double quotes ( " ) for certain special signals, such as looking for environment variable names to evaluate. Read more about Quoting in the shell.)
So what does ^> mean to grep? We know that contig name lines always start with a > character, so > is a literal for grep to use in its pattern match.
We might be able to get away with just using this literal alone as our regex, specifying '>' as the command line argument. But for grep, the more specific the pattern, the better. So we constrain where the > can appear on the line. The special carat ( ^ ) metacharacter represents "beginning of line". So ^> means "beginning of a line followed by a > character".
Exercise: How many contigs are there in the sacCer3 reference?
| Expand |
|---|
|
| Code Block |
|---|
| language | bash |
|---|
| title | Get the alignment exercises files |
|---|
| mkdir -p $SCRATCH/core_ngs/references/fasta
cp $CORENGS/references/*.fa $SCRATCH/core_ngs/references/fasta/ |
|
| Expand |
|---|
|
| Code Block |
|---|
| cd $SCRATCH/core_ngs/references/fasta
grep -P '^>' sacCer3.fa | wc -l |
Or use grep's -c option that says "just count the line matches" | Code Block |
|---|
| grep -P -c '^>' sacCer3.fa |
|
| Expand |
|---|
|
There are 17 contigs. |
Aligner overview
There are many aligners available, but we will concentrate on two of the most popular general-purpose ones: bwa and bowtie2. The table below outlines the available protocols for them.
| alignment type | aligner options | pro's | con's |
|---|
| global with bwa | SE: PE: - bwa aln <R1>
- bwa aln <R2>
- bwa sampe
| - simple to use (take default options)
- good for basic global alignment
| |
| global with bowtie2 | bowtie2 --global | - extremely configurable
- can be used for RNAseq alignment (after adapter trimming) because of its many options
| |
| local with bwa | bwa mem | - simple to use (take default options)
- very fast
- no adapter trimming needed
- good for simple RNAseq analysis
- the secondary alignments it reports provide splice junction information
| - always produces alignments with secondary reads
- must be filtered if not desired
|
| local with bowtie2 | bowtie2 --local | - extremely configurable
- no adapter trimming needed
- good for small RNA alignment because of its many options
| |
Exercise #1: BWA global alignment – Yeast ChIP-seq
Overview ChIP-seq alignment workflow with BWA
We will perform a global alignment of the paired-end Yeast ChIP-seq sequences using bwa. This workflow has the following steps:
- Trim the FASTQ sequences down to 50 with fastx_clipper
- this removes most of any 5' adapter contamination without the fuss of specific adapter trimming w/cutadapt
- Prepare the sacCer3 reference index for bwa using bwa index
- this is done once, and re-used for later alignments
- Perform a global bwa alignment on the R1 reads (bwa aln) producing a BWA-specific binary .sai intermediate file
- Perform a global bwa alignment on the R2 reads (bwa aln) producing a BWA-specific binary .sai intermediate file
- Perform pairing of the separately aligned reads and report the alignments in SAM format using bwa sampe
- Convert the SAM file to a BAM file (samtools view)
- Sort the BAM file by genomic location (samtools sort)
- Index the BAM file (samtools index)
- Gather simple alignment statistics (samtools flagstat and samtools idxstat)
We're going to skip the trimming step for now and see how it goes. We'll perform steps 2 - 5 now and leave samtools for a later exercise since steps 6 - 10 are common to nearly all post-alignment workflows.
Introducing BWA
Like other tools you've worked with so far, you first need to load bwa. Do that now, and then enter bwa with no arguments to view the top-level help page (many NGS tools will provide some help when called with no arguments). Note that bwa is available both from the standard TACC module system and as BioContainers. module.
| Expand |
|---|
| title | Make sure you're in a idev session |
|---|
|
| Code Block |
|---|
| language | bash |
|---|
| title | Start an idev session |
|---|
| idev -p normal -m 120 -A UT-2015-05-18 -N 1 -n 68 |
|
| Code Block |
|---|
|
module load biocontainers # takes a while
module load bwa
bwa |
| Code Block |
|---|
|
Program: bwa (alignment via Burrows-Wheeler transformation)
Version: 0.7.17-r1188
Contact: Heng Li <lh3@sanger.ac.uk>
Usage: bwa <command> [options]
Command: index index sequences in the FASTA format
mem BWA-MEM algorithm
fastmap identify super-maximal exact matches
pemerge merge overlapping paired ends (EXPERIMENTAL)
aln gapped/ungapped alignment
samse generate alignment (single ended)
sampe generate alignment (paired ended)
bwasw BWA-SW for long queries
shm manage indices in shared memory
fa2pac convert FASTA to PAC format
pac2bwt generate BWT from PAC
pac2bwtgen alternative algorithm for generating BWT
bwtupdate update .bwt to the new format
bwt2sa generate SA from BWT and Occ
Note: To use BWA, you need to first index the genome with `bwa index'.
There are three alignment algorithms in BWA: `mem', `bwasw', and
`aln/samse/sampe'. If you are not sure which to use, try `bwa mem'
first. Please `man ./bwa.1' for the manual. |
As you can see, bwa include many sub-commands that perform the tasks we are interested in.
Building the BWA sacCer3 index
We will index the genome with the bwa index command. Type bwa index with no arguments to see usage for this sub-command.
| Code Block |
|---|
|
Usage: bwa index [options] <in.fasta>
Options: -a STR BWT construction algorithm: bwtsw, is or rb2 [auto]
-p STR prefix of the index [same as fasta name]
-b INT block size for the bwtsw algorithm (effective with -a bwtsw) [10000000]
-6 index files named as <in.fasta>.64.* instead of <in.fasta>.*
Warning: `-a bwtsw' does not work for short genomes, while `-a is' and
`-a div' do not work not for long genomes. |
Based on the usage description, we only need to specify two things:
- The name of the FASTA file
- Whether to use the bwtsw or is algorithm for indexing
Since sacCer3 is relative large (~12 Mbp) we will specify bwtsw as the indexing option (as indicated by the "Warning" message), and the name of the FASTA file is sacCer3.fa.
The output of this command is a group of files that are all required together as the index. So, within our references directory, we will create another directory called references/bwa/sacCer3 and build the index there. To remind ourselves which FASTA was used to build the index, we create a symbolic link to our references/fasta/sacCer3.fa file (note the use of the ../.. relative path syntax).
| Expand |
|---|
|
| Code Block |
|---|
| language | bash |
|---|
| title | Get the alignment exercises files |
|---|
|
|
| Code Block |
|---|
|
grep -P '^>' sacCer3.fa | wc -l |
Or use grep's -c option that says "just count the line matches"
| Code Block |
|---|
|
grep -P -c '^>' sacCer3.fa |
| Expand |
|---|
|
There are 17 contigs. |
Performing the bwa alignment
Now, we're ready to execute the actual alignment, with the goal of initially producing a SAM file from the input FASTQ files and reference. First prepare a directory for this exercise and link the sacCer3 reference directories there (this will make our commands more readable).
| Code Block |
|---|
|
| language | bash |
|---|
title | Prepare to align yeast data | mkdir -p $SCRATCH/core_ngs/alignment/ |
yeast_bwa
cdfastq
mkdir -p $SCRATCH/core_ngs/ |
alignmentyeast_bwaln -s -f ../fastq
ln -s -f ../../references/bwa/sacCer3 |
As our workflow indicated, we first use bwa aln on the R1 and R2 FASTQs, producing a BWA-specific .sai intermediate binary files.
What does bwa aln needs in the way of arguments?
cp $CORENGS/alignment/*fastq.gz $SCRATCH/core_ngs/alignment/fastq/
cp $CORENGS/references/*.fa $SCRATCH/core_ngs/references/fasta/ |
|
| Code Block |
|---|
| language | bash |
|---|
| title | Prepare BWA reference directory for sacCer3 |
|---|
|
mkdir -p $SCRATCH/core_ngs/references/bwa/sacCer3
cd $SCRATCH/core_ngs/references/bwa/sacCer3
ln -s ../../fasta/sacCer3.fa
ls -l |
Now execute the bwa index command.
There are lots of options, but here is a summary of the most important ones.
...
| title | Build BWA index for sacCer3 |
|---|
|
bwa index -a bwtsw sacCer3.fa |
Since the yeast genome is not large when compared to human, this should not take long to execute (otherwise we would do it as a batch job). When it is complete you should see a set of index files like this:
| Code Block |
|---|
| title | BWA index files for sacCer3 |
|---|
|
sacCer3.fa
sacCer3.fa.amb
sacCer3.fa.ann
sacCer3.fa.bwt
sacCer3.fa.pac
sacCer3.fa.sa |
Performing the bwa alignment
Now, we're ready to execute the actual alignment, with the goal of initially producing a SAM file from the input FASTQ files and reference. First prepare a directory for this exercise and link the sacCer3 reference directories there (this will make our commands more readable).
| Expand |
|---|
|
| Code Block |
|---|
| # Copy the pre-built references
mkdir -p $SCRATCH/core_ngs/references
cp $CORENGS/references/*.fa $SCRATCH/core_ngs/references/fasta/
# Get the FASTQ to align
mkdir -p $SCRATCH/core_ngs/alignment/fastq
cp $CORENGS/alignment/*fastq.gz $SCRATCH/core_ngs/alignment/fastq/
|
|
Other options control the details of how much a mismatch or gap is penalized, limits on the number of acceptable hits per read, and so on. Much more information can be found on the BWA manual page.
For a basic alignment like this, we can just go with the default alignment parameters.
Also note that bwa writes its (binary) output to standard output by default, so we need to redirect that to a .sai file.
We will execute these commands directly (not in a batch job), but since they are fairly large files we will first set up an interactive development (idev) session, which will give us a compute node for a couple of hours:
| Code Block |
|---|
| language | bash |
|---|
| title | Start an idev session |
|---|
|
idev -p normal -m 120 -N 1 -n 24 -A UT-2015-05-18 --reservation=CCBB |
| Tip |
|---|
You can tell you're in a idev session because the hostname command will return a compute node name (e.g. nid00438) instead of a login node name (e.g. login5). |
...
| Code Block |
|---|
| language | bash |
|---|
| title | bwa aln commands for yeast R1 and R2 | Prepare to align yeast data |
|---|
|
mkdir -p $SCRATCH/core_ngs/alignment/yeast_module load bwa
cd $SCRATCH/core_ngs/alignment/yeast_bwa
bwaln aln sacCer3/sacCer3.fa fastq/Sample_Yeast_L005_R1.cat.fastq.gz > yeast_R1.sai
bwa aln sacCer3/sacCer3.fa fastq/Sample_Yeast_L005_R2.cat.fastq.gz > yeast_R2.sai |
When all is done you should have two .sai files: yeast_R1.sai and yeast_R2.sai. Double check that output was written by doing ls -lh and making sure the file sizes listed are not 0.
Exercise: How long did it take to align the R2 file?
-s -f ../fastq
ln -s -f ../../references/bwa/sacCer3 |
As our workflow indicated, we first use bwa aln on the R1 and R2 FASTQs, producing a BWA-specific .sai intermediate binary files.
What does bwa aln needs in the way of arguments?
| Expand |
|---|
|
| Expand |
|---|
|
The last few lines of bwa's execution output should look something like this: |
| Code Block |
|---|
| [bwa_aln_core] write to the disk... 0.01 sec
[bwa_aln_core] 592180 sequences have been processed.
[main] Version: 0.7.12-r1039
[main] CMD: bwa aln sacCer3/sacCer3.fa fastq/Sample_Yeast_L005_R2.cat.fastq.gz
[main] Real time: 218.832 sec; CPU: 218.274 sec |
So the R2 alignment took just under 4 minutes. |
Since you have your own private compute node, you can use all its resources. It has 24 cores, so re-run the R2 alignment asking for 20 execution threads.
| Code Block |
|---|
bwa aln -t 20 sacCer3/sacCer3.fa fastq/Sample_Yeast_L005_R2.cat.fastq.gz > yeast_R2.sai |
Exercise: How much of a speedup did you seen when aligning the R2 file with 20 threads?
There are lots of options, but here is a summary of the most important ones.
| Option | Effect |
|---|
| -l | Specifies the length of the seed (default = 32) |
| -k | Specifies the number of mismatches allowable in the seed of each alignment (default = 2) |
| -n | Specifies the number of mismatches (or fraction of bases in a given alignment that can be mismatches) in the entire alignment (including the seed) (default = 0.04) |
| -t | Specifies the number of threads |
Other options control the details of how much a mismatch or gap is penalized, limits on the number of acceptable hits per read, and so on. Much more information can be found on the BWA manual page.
For a basic alignment like this, we can just go with the default alignment parameters.
Note that bwa writes its (binary) output to standard output by default, so we need to redirect that to a .sai file.
For simplicity, we will just execute these commands directly, one at a time. Each command should only take few minutes and you will see bwa's progress messages in your terminal.
| Code Block |
|---|
| language | bash |
|---|
| title | bwa aln commands for yeast R1 and R2 |
|---|
|
# If not already loaded:
module load biocontainers
module load bwa
cd $SCRATCH/core_ngs/alignment/yeast_bwa
bwa aln |
| Expand |
|---|
|
The last few lines of bwa's execution output should look something like this: | Code Block |
|---|
|
|
[bwa_aln_core] write to the disk... 0.01 sec
[bwa_aln_core] 592180 sequences have been processed.
[main] Version: 0.7.12-r1039
[main] CMD: bwa aln -t 20 sacCer3/sacCer3.fa fastq/Sample_Yeast_L005_
R2R1.cat.fastq.gz
[main]
Real time: 21.157 sec; CPU: 268.813 secSo the R2 alignment took only 21 seconds, or about 10 times as fast as with only one processing thread.
Next we use the bwa sampe command to pair the reads and output SAM format data. Just type that command in with no arguments to see its usage.
For this command you provide the same reference index prefix as for bwa aln, along with the two .sai files and the two original FASTQ files. Also, bwa writes its output to standard output, so redirect that to a .sam file.
...
> yeast_R1.sai
bwa aln sacCer3/sacCer3.fa fastq/Sample_Yeast_L005_R2.cat.fastq.gz > yeast_R2.sai |
When all is done you should have two .sai files: yeast_R1.sai and yeast_R2.sai.
| Tip |
|---|
| title | Make sure your output files are not empty |
|---|
|
Double check that output was written by doing ls -lh and making sure the file sizes listed are not 0. |
Exercise: How long did it take to align the R2 file?
| Expand |
|---|
|
The last few lines of bwa's execution output should look something like this: |
This looks for the pattern '^HWI' which is the start of every read name (which starts every alignment record).
Remember -c says just count the records, don't display them.
| Code Block |
|---|
|
grep -P -c '^HWI' yeast_pairedend.sam |
Or use the -v (invert) option to tell grep to print all lines that don't match a particular pattern; here, all the header lines that start with @.
| Code Block |
|---|
|
grep -P -v -c '^@' yeast_pairedend.sam |
| Expand |
|---|
|
| There are 1,184,360 alignment records. |
Exercise: How many sequences were in the R1 and R2 FASTQ files combined?
| Expand |
|---|
|
| zcat |
| title | BWA global alignment of R1 reads |
|---|
bwa sampe sacCer3/sacCer3.fa yeast_R1.sai yeast_R2.sai fastq/Sample_Yeast_L005_R1.cat.fastq.gz fastq/Sample_Yeast_L005_R2.cat.fastq.gz > yeast_pairedend.sam |
You should now have a SAM file (yeast_pairedend.sam) that contains the alignments. It's just a text file, so take a look with head, more, less, tail, or whatever you feel like. In the next section, with samtools, you'll learn some additional ways to analyze the data once you create a BAM file.
Exercise: What kind of information is in the first lines of the SAM file?
| Expand |
|---|
|
| The SAM file has a number of header lines, which all start with an at sign ( @ ). The @SQ lines describe each contig and its length. There is also a @PG line that describes the way the bwa sampe was performed. |
Exercise: How many alignment records (not header records) are in the SAM file?
...
[bwa_aln] 17bp reads: max_diff = 2
[bwa_aln] 38bp reads: max_diff = 3
[bwa_aln] 64bp reads: max_diff = 4
[bwa_aln] 93bp reads: max_diff = 5
[bwa_aln] 124bp reads: max_diff = 6
[bwa_aln] 157bp reads: max_diff = 7
[bwa_aln] 190bp reads: max_diff = 8
[bwa_aln] 225bp reads: max_diff = 9
[bwa_aln_core] calculate SA coordinate... 50.76 sec
[bwa_aln_core] write to the disk... 0.07 sec
[bwa_aln_core] 262144 sequences have been processed.
[bwa_aln_core] calculate SA coordinate... 50.35 sec
[bwa_aln_core] write to the disk... 0.07 sec
[bwa_aln_core] 524288 sequences have been processed.
[bwa_aln_core] calculate SA coordinate... 13.64 sec
[bwa_aln_core] write to the disk... 0.01 sec
[bwa_aln_core] 592180 sequences have been processed.
[main] Version: 0.7.17-r1188
[main] CMD: /usr/local/bin/bwa aln sacCer3/sacCer3.fa fastq/Sample_Yeast_L005_ |
R[12] | wc -l | awk '{print $1/4}'
|
| Expand |
|---|
|
| There were a total of 1,184,360 original sequences |
Exercises:
...
[main] Real time: 122.936 sec; CPU: 123.597 sec |
So the R2 alignment took ~123 seconds (~2 minutes). |
Since you have your own private compute node, you can use all its resources. It has 68 cores, so re-run the R2 alignment asking for 60 execution threads.
| Code Block |
|---|
bwa aln -t 60 sacCer3/sacCer3.fa fastq/Sample_Yeast_L005_R2.cat.fastq.gz > yeast_R2.sai |
Exercise: How much of a speedup did you seen when aligning the R2 file with 20 threads
...
?
| Expand |
|---|
|
Both R1 and R2 reads must have separate alignment records, because there were 1,184,360 R1+R2 reads and the same number of alignment records. The SAM file must contain both mapped and un-mapped reads, because there were 1,184,360 R1+R2 reads and the same number of alignment records. Alignment records occur in the same read-name order as they did in the FASTQ, except that they come in pairs. The R1 read comes 1st, then the corresponding R2. This is called read name ordering. |
Using cut to isolate fields
Recall the format of a SAM alignment record:
 Image Removed
Image Removed
Suppose you wanted to look only at field 3 (contig name) values in the SAM file. You can do this with the handy cut command. Below is a simple example where you're asking cut to display the 3rd column value for the last 10 alignment records.
| Code Block |
|---|
| language | bash |
|---|
| title | Cut syntax for a single field |
|---|
|
tail yeast_pairedend.sam | cut -f 3 |
By default cut assumes the field delimiter is Tab, which is the delimiter used in the majority of NGS file formats. You can specify a different delimiter with the -d option.
You can also specify a range of fields, and mix adjacent and non-adjacent fields. This displays fields 2 through 6, field 9, and all fields starting with the 12th (SAM tag fields).
| Code Block |
|---|
| language | bash |
|---|
| title | Cut syntax for multiple fields |
|---|
|
tail yeast_pairedend.sam | cut -f 2-6,9,12- |
You may have noticed that some alignment records contain contig names (e.g. chrV) in field 3 while others contain an asterisk ( * ). Usually the * means the record didn't map. (This isn't always true – later you'll see how to properly distinguish between mapped and unmapped reads using samtools.) We're going to use this heuristic along with cut to see about how many records represent aligned sequences.
First we need to make sure that we don't look at fields in the SAM header lines. We're going to end up with a series of pipe operations, and the best way to make sure you're on track is to enter them one at a time piping to head:
| Code Block |
|---|
| language | bash |
|---|
| title | Grep pattern that doesn't match header |
|---|
|
# the ^HWI pattern matches lines starting with HWI (the start of all read names in column 1)
grep -P '^HWI' yeast_pairedend.sam | head |
Ok, it looks like we're seeing only alignment records. Now let's pull out only field 3 using cut:
| Code Block |
|---|
| language | bash |
|---|
| title | Get contig name info with cut |
|---|
|
grep -P '^HWI' yeast_pairedend.sam | cut -f 3 | head |
Cool, we're only seeing the contig name info now. Next we use grep again, piping it our contig info and using the -v (invert) switch to say print lines that don't match the pattern:
| Code Block |
|---|
| language | bash |
|---|
| title | Filter contig name of * (unaligned) |
|---|
|
grep -P '^HWI' yeast_pairedend.sam | cut -f 3 | grep -v '*' | head |
Perfect! We're only seeing real contig names that (usually) represent aligned reads. Let's count them by piping to wc -l (and omitting omit head of course – we want to count everything).
| Code Block |
|---|
| language | bash |
|---|
| title | Count aligned SAM records |
|---|
|
grep -P '^HWI' yeast_pairedend.sam | cut -f 3 | grep -v '*' | wc -l |
Exercise: About how many records represent aligned sequences? What alignment rate does this represent?
| Expand |
|---|
|
The expression above returns 612,968. There were 1,184,360 records total, so the percentage is: | Code Block |
|---|
| language | bash |
|---|
| title | Calculate alignment rate |
|---|
| echo $((612968 * 100/ 1184360)) |
or about 51%. Not great. |
Exercise: What might we try in order to improve the alignment rate?
| Expand |
|---|
|
| Recall that these are 100 bp reads and we did not remove adapter contamination. There will be a distribution of fragment sizes – some will be short – and those short fragments may not align without adapter removal (e.g. with fastx_trimmer). |
Exercise #2: Simple SAMtools Utilities
We have used several alignment methods that all generate results in the form of the near-universal SAM/BAM file format. The SAMtools program is a ubiquitously used set of tools that allow a user to manipulate SAM/BAM files in many different ways, ranging from simple tasks (like SAM/BAM interconversion) to more complex functions (like removal of PCR duplicates). It is available in the TACC module system in the typical fashion.
In this exercise, we will use five very simple utilities provided by samtools: view, sort, index, flagstat, and idxstats. Each of these is executed in one line for a given SAM/BAM file. In the SAMtools/BEDtools section tomorrow we will explore samtools in more in depth.
For the sake of time and simplicity, here we are only going to run these commands on the yeast paired-end alignment file. The same commands can be run on the other files by changing the names, so feel free to try them on other SAM files. Indeed, it is very common in practice to use bash loops to generate many commands for a large set of alignments and deposit those commands into a batch job cmds file for submission.
To start, we will move to the directory containing our SAM files, among other things, and load up samtools using the module system. After loading it, just run the samtools command to see what the available tools are (and to see what the syntax of an actual command is).
| Code Block |
|---|
|
cd $SCRATCH/core_ngs/alignment
ls -la
module load samtools
samtools |
You will see the following screen after running samtools with no other options:
| Code Block |
|---|
Program: samtools (Tools for alignments in the SAM format)
Version: 1.2 (using htslib 1.2.1)
Usage: samtools <command> [options]
Commands:
-- indexing
faidx index/extract FASTA
index index alignment
-- editing
calmd recalculate MD/NM tags and '=' bases
fixmate fix mate information
reheader replace BAM header
rmdup remove PCR duplicates
targetcut cut fosmid regions (for fosmid pool only)
-- file operations
bamshuf shuffle and group alignments by name
cat concatenate BAMs
merge merge sorted alignments
mpileup multi-way pileup
sort sort alignment file
split splits a file by read group
bam2fq converts a BAM to a FASTQ
-- stats
bedcov read depth per BED region
depth compute the depth
flagstat simple stats
idxstats BAM index stats
phase phase heterozygotes
stats generate stats (former bamcheck)
-- viewing
flags explain BAM flags
tview text alignment viewer
view SAM<->BAM<->CRAM conversion |
| Tip |
|---|
| title | SAMtools version differences |
|---|
|
Be sure to check what version of samtools you are using! The most recent edition of SAMtools is 1.2, which has some important differences from the last version, 0.1.19. Most commands for this section are the same between the two versions, but if you see code from other sources using samtools, the version differences may be important. |
Samtools view
The utility samtools view provides a way of converting SAM (text format) files to BAM (binary, compressed) files directly. It also provides many, many other functions which we will discuss lster. To get a preview, execute samtools view without any other arguments. You should see:
| Code Block |
|---|
Usage: samtools view [options] <in.bam>|<in.sam>|<in.cram> [region ...]
Options: -b output BAM
-C output CRAM (requires -T)
-1 use fast BAM compression (implies -b)
-u uncompressed BAM output (implies -b)
-h include header in SAM output
-H print SAM header only (no alignments)
-c print only the count of matching records
-o FILE output file name [stdout]
-U FILE output reads not selected by filters to FILE [null]
-t FILE FILE listing reference names and lengths (see long help) [null]
-T FILE reference sequence FASTA FILE [null]
-L FILE only include reads overlapping this BED FILE [null]
-r STR only include reads in read group STR [null]
-R FILE only include reads with read group listed in FILE [null]
-q INT only include reads with mapping quality >= INT [0]
-l STR only include reads in library STR [null]
-m INT only include reads with number of CIGAR operations
consuming query sequence >= INT [0]
-f INT only include reads with all bits set in INT set in FLAG [0]
-F INT only include reads with none of the bits set in INT
set in FLAG [0]
-x STR read tag to strip (repeatable) [null]
-B collapse the backward CIGAR operation
-s FLOAT integer part sets seed of random number generator [0];
rest sets fraction of templates to subsample [no subsampling]
-@ INT number of BAM compression threads [0]
-? print long help, including note about region specification
-S ignored (input format is auto-detected) |
That is a lot to process! For now, we just want to read in a SAM file and output a BAM file. The input format is auto-detected, so we don't need to say that we're inputing a SAM instead of a BAM. We just need to tell the tool to output the file in BAM format, and provide the name of the destination BAM file. This command is as follows:
| Code Block |
|---|
|
samtools view -b yeast_pairedend.sam -o yeast_pairedend.bam |
- the -b option tells the tool to output BAM format
- the -o option specifies the name of the output BAM file that will be created
How do you look at the BAM file contents now? That's simple. Just use samtools view. without the -b option. Remember to pipe output to a pager!
| Code Block |
|---|
|
samtools view yeast_pairedend.bam | more
|
Samtools sort
Look at the SAM file briefly using less. You will notice, if you scroll down, that the alignments are in no particular order, with chromosomes and start positions all mixed up. This makes searching through the file very inefficient. samtools sort is a piece of samtools that provides the ability to re-order entries in the SAM file either by coordinate position or by read name.
If you execute samtools sort without any options, you see its help page:
| Code Block |
|---|
Usage: samtools sort [options...] [in.bam]
Options:
-l INT Set compression level, from 0 (uncompressed) to 9 (best)
-m INT Set maximum memory per thread; suffix K/M/G recognized [768M]
-n Sort by read name
-o FILE Write final output to FILE rather than standard output
-O FORMAT Write output as FORMAT ('sam'/'bam'/'cram') (either -O or
-T PREFIX Write temporary files to PREFIX.nnnn.bam -T is required)
-@ INT Set number of sorting and compression threads [1]
Legacy usage: samtools sort [options...] <in.bam> <out.prefix>
Options:
-f Use <out.prefix> as full final filename rather than prefix
-o Write final output to stdout rather than <out.prefix>.bam
-l,m,n,@ Similar to corresponding options above |
In most cases you will be sorting a BAM file by position rather than by name. You can use either -o or reidrection with > to control the output.
To sort the paired-end yeast BAM file by coordinate, and get a BAM file named yeast_pairedend.sort.bam as output, execute the following command:
| Code Block |
|---|
|
samtools sort -O bam -T yeast_pairedend.sort yeast_pairedend.bam > yeast_pairedend.sort.bam |
- The -O options says the output format should be BAM
- The -T options gives a prefix for temporary files produced during sorting
- By default sort writes its output to standard output, so we use > to redirect to a file named yeast_pairedend.sort.bam
Samtools index
Many tools (like the UCSC Genome Browser) only need to use sub-sections of the BAM file at a given point in time. For example, if you are viewing alignments that are within a particular gene alignments on other chromosomes generally do not need to be loaded. In order to speed up access, BAM files are indexed, producing BAI files which allow other programs to navigate directly to the alignments of interest. This is especially important when you have many alignments.
The utility samtools index creates an index that has the exact name as the input BAM file, with suffix .bai appended. The help page, if you execute samtools index with no arguments, is as follows:
| Code Block |
|---|
Usage: samtools index [-bc] [-m INT] <in.bam> [out.index]
Options:
-b Generate BAI-format index for BAM files [default]
-c Generate CSI-format index for BAM files
-m INT Set minimum interval size for CSI indices to 2^INT [14] |
Here, the syntax is way, way easier. We want a BAI-format index which is the default. (CSI-format is used with extremely long contigs, which we aren't considering here - the most common use case are highly polyploid plant genomes).
So all we have to type is:
| Code Block |
|---|
|
samtools index yeast_pairedend.sort.bam |
This will produce a file named yeast_pairedend.bam.bai.
Most of the time when an index is required, it will be automatically located as long as it is in the same directory as its BAM file and shares the same name up until the .bai extension.
Samtools idxstats
Now that we have a sorted, indexed BAM file, we might like to get some simple statistics about the alignment run. For example, we might like to know how many reads aligned to each chromosome/contig. The samtools idxstats is a very simple tool that provides this information. If you type the command without any arguments, you will see that it could not be simpler - just type the following command:
| Code Block |
|---|
|
samtools idxstats yeast_pairedend.sort.bam |
The output is a text file with four tab-delimited columns with the following meanings:
- chromosome name
- chromosome length
- number of mapped reads
- number of unmapped reads
The reason that the "unmapped reads" field for the named chromosomes is not zero is that, if one half of a pair of reads aligns while the other half does not, the unmapped read is still assigned to the chromosome its mate mapped to, but is flagged as unmapped.
| Tip |
|---|
If you're mapping to a non-genomic reference such as miRBase miRNAs or another set of genes (a transcriptome), samtools idxstats gives you a quick look at quantitative alignment results. |
Samtools flagstat
Finally, we might like to obtain some other statistics, such as the percent of all reads that aligned to the genome. The samtools flagstat tool provides very simple analysis of the SAM flag fields, which includes information like whether reads are properly paired, aligned or not, and a few other things. Its syntax is identical to that of samtools idxstats:
| Code Block |
|---|
|
samtools flagstat yeast_pairedend.sort.bam |
You should see something like this:
| Code Block |
|---|
1184360 + 0 in total (QC-passed reads + QC-failed reads)
0 + 0 secondary
0 + 0 supplementary
0 + 0 duplicates
547664 + 0 mapped (46.24%:-nan%)
1184360 + 0 paired in sequencing
592180 + 0 read1
592180 + 0 read2
473114 + 0 properly paired (39.95%:-nan%)
482360 + 0 with itself and mate mapped
65304 + 0 singletons (5.51%:-nan%)
534 + 0 with mate mapped to a different chr
227 + 0 with mate mapped to a different chr (mapQ>=5) |
Ignore the "+ 0" addition to each line - that is a carry-over convention for counting QA-failed reads that is no longer necessary.
The most important statistic is the mapping rate, but this readout allows you to verify that some common expectations (e.g. that about the same number of R1 and R2 reads aligned, and that most mapped reads are proper pairs) are met.
Exercise #6: Yeast BWA PE alignment with Anna's script
Now that you've done everything the hard way, let's see how to do run an alignment pipeline using Anna's script.
First the setup:
|
The last few lines of bwa's execution output should look something like this: | Code Block |
|---|
| [bwa_aln] 17bp reads: max_diff = 2
[bwa_aln] 38bp reads: max_diff = 3
[bwa_aln] 64bp reads: max_diff = 4
[bwa_aln] 93bp reads: max_diff = 5
[bwa_aln] 124bp reads: max_diff = 6
[bwa_aln] 157bp reads: max_diff = 7
[bwa_aln] 190bp reads: max_diff = 8
[bwa_aln] 225bp reads: max_diff = 9
[bwa_aln_core] calculate SA coordinate... 266.70 sec
[bwa_aln_core] write to the disk... 0.04 sec
[bwa_aln_core] 262144 sequences have been processed.
[bwa_aln_core] calculate SA coordinate... 268.94 sec
[bwa_aln_core] write to the disk... 0.03 sec
[bwa_aln_core] 524288 sequences have been processed.
[bwa_aln_core] calculate SA coordinate... 72.26 sec
[bwa_aln_core] write to the disk... 0.01 sec
[bwa_aln_core] 592180 sequences have been processed.
[main] Version: 0.7.17-r1188
[main] CMD: /usr/local/bin/bwa aln -t 60 sacCer3/sacCer3.fa fastq/Sample_Yeast_L005_R2.cat.fastq.gz
[main] Real time: 19.872 sec; CPU: 617.095 sec |
So the R2 alignment took only ~20 seconds (real time), or 6+ times as fast as with only one processing thread. Note, though, that the CPU time with 60 threads was greater (617 sec) than with only 1 thread (124 sec). That's because of the thread management overhead when using multiple threads. |
Next we use the bwa sampe command to pair the reads and output SAM format data. Just type that command in with no arguments to see its usage.
For this command you provide the same reference index prefix as for bwa aln, along with the two .sai files and the two original FASTQ files. Also, bwa writes its output to standard output, so redirect that to a .sam file.
Here is the command line statement you need. Just execute it on the command line.
| Code Block |
|---|
| language | bash |
|---|
| title | Pairing of BWA R1 and R2 aligned reads |
|---|
|
bwa sampe sacCer3/sacCer3.fa yeast_R1.sai yeast_R2.sai \
fastq/Sample_Yeast_L005_R1.cat.fastq.gz \
fastq/Sample_Yeast_L005_R2.cat.fastq.gz > yeast_pairedend.sam |
You should now have a SAM file (yeast_pairedend.sam) that contains the alignments. It's just a text file, so take a look with head, more, less, tail, or whatever you feel like. Later you'll learn additional ways to analyze the data with samtools once you create a BAM file.
Exercise: What kind of information is in the first lines of the SAM file?
| Expand |
|---|
|
The SAM file has a number of header lines, which all start with an at sign ( @ ). The @SQ lines describe each contig (chromosome) and its length. There is also a @PG line that describes the way the bwa sampe was performed. |
Exercise: How many alignment records (not header records) are in the SAM file?
| Expand |
|---|
|
This looks for the pattern '^HWI' which is the start of every read name (which starts every alignment record).
Remember -c says just count the records, don't display them. | Code Block |
|---|
| grep -P -c '^HWI' yeast_pairedend.sam |
Or use the -v (invert) option to tell grep to print all lines that don't match a particular pattern; here, all header lines, which start with @. | Code Block |
|---|
| grep -P -v -c '^@' yeast_pairedend.sam |
|
| Expand |
|---|
|
| There are 1,184,360 alignment records. |
Exercise: How many sequences were in the R1 and R2 FASTQ files combined?
| Expand |
|---|
|
zcat fastq/Sample_Yeast_L005_R[12].cat.fastq.gz | wc -l | awk '{print $1/4}'
|
| Expand |
|---|
|
| There were a total of 1,184,360 original sequences (R1s + R2s) |
Exercises:
- Do both R1 and R2 reads have separate alignment records?
- Does the SAM file contain both mapped and un-mapped reads?
- What is the order of the alignment records in this SAM file?
| Expand |
|---|
|
Both R1 and R2 reads must have separate alignment records, because there were 1,184,360 R1+R2 reads and the same number of alignment records. The SAM file must contain both mapped and un-mapped reads, because there were 1,184,360 R1+R2 reads and the same number of alignment records. Alignment records occur in the same read-name order as they did in the FASTQ, except that they come in pairs. The R1 read comes 1st, then the corresponding R2. This is called read name ordering. |
Using cut to isolate fields
Recall the format of a SAM alignment record:
 Image Added
Image Added
Suppose you wanted to look only at field 3 (contig name) values in the SAM file. You can do this with the handy cut command. Below is a simple example where you're asking cut to display the 3rd column value for the last 10 alignment records.
| Code Block |
|---|
| language | bash |
|---|
| title | Cut syntax for a single field |
|---|
|
tail yeast_pairedend.sam | cut -f 3 |
By default cut assumes the field delimiter is Tab, which is the delimiter used in the majority of NGS file formats. You can specify a different delimiter with the -d option.
You can also specify a range of fields, and mix adjacent and non-adjacent fields. This displays fields 2 through 6, field 9:
| Code Block |
|---|
| language | bash |
|---|
| title | Cut syntax for multiple fields |
|---|
|
tail -20 yeast_pairedend.sam | cut -f 2-6,9 |
You may have noticed that some alignment records contain contig names (e.g. chrV) in field 3 while others contain an asterisk ( * ). The * means the record didn't map. We're going to use this heuristic along with cut to see about how many records represent aligned sequences. (Note this is not the strictly correct method of finding unmapped reads because not all unmapped reads have an asterisk in field 3. Later you'll see how to properly distinguish between mapped and unmapped reads using samtools.)
First we need to make sure that we don't look at fields in the SAM header lines. We're going to end up with a series of pipe operations, and the best way to make sure you're on track is to enter them one at a time piping to head:
| Code Block |
|---|
| language | bash |
|---|
| title | Grep pattern that doesn't match header |
|---|
|
# the ^@ pattern matches lines starting with @ (only header lines),
# and -v says output lines that don't match
grep -v -P '^@' yeast_pairedend.sam | head |
Ok, it looks like we're seeing only alignment records. Now let's pull out only field 3 using cut:
| Code Block |
|---|
| language | bash |
|---|
| title | Get contig name info with cut |
|---|
|
grep -v -P '^@' yeast_pairedend.sam | cut -f 3 | head |
Cool, we're only seeing the contig name info now. Next we use grep again, piping it our contig info and using the -v (invert) switch to say print lines that don't match the pattern:
| Code Block |
|---|
| language | bash |
|---|
| title | Filter contig name of * (unaligned) |
|---|
|
grep -v -P '^@' yeast_pairedend.sam | cut -f 3 | grep -v '*' | head |
Perfect! We're only seeing real contig names that (usually) represent aligned reads. Let's count them by piping to wc -l (and omitting omit head of course – we want to count everything).
| Code Block |
|---|
| language | bash |
|---|
| title | Count aligned SAM records |
|---|
|
grep -v -P '^@' yeast_pairedend.sam | cut -f 3 | grep -v '*' | wc -l |
Exercise: About how many records represent aligned sequences? What alignment rate does this represent?
| Expand |
|---|
|
The expression above returns 612,968. There were 1,184,360 records total, so the percentage is: | Code Block |
|---|
| language | bash |
|---|
| title | Calculate alignment rate |
|---|
| awk 'BEGIN{print 612968/1184360}' |
or about 51%. Not great. Note we perform this calculation in awk's BEGIN block, which is always executed, instead of the body block, which is only executed for lines of input. And here we call awk without piping it any input. See Linux fundamentals: cut,sort,uniq,grep,awk |
Exercise: What might we try in order to improve the alignment rate?
| Expand |
|---|
|
| Recall that these are 100 bp reads and we did not remove adapter contamination. There will be a distribution of fragment sizes – some will be short – and those short fragments may not align without adapter removal (e.g. with fastx_trimmer). |
Exercise #2: Basic SAMtools Utilities
The SAMtools program is a commonly used set of tools that allow a user to manipulate SAM/BAM files in many different ways, ranging from simple tasks (like SAM/BAM format conversion) to more complex functions (like sorting, indexing and statistics gathering). It is available in the TACC module system (as well as in BioContainers). Load that module and see what samtools has to offer:
| Expand |
|---|
| title | Make sure you're in a idev session |
|---|
|
| Code Block |
|---|
| language | bash |
|---|
| title | Start an idev session |
|---|
| idev -p normal -m 120 -A UT-2015-05-18 -N 1 -n 68 |
|
| Code Block |
|---|
|
# If not already loaded
module load biocontainers # takes a while
module load samtools
samtools |
| Code Block |
|---|
| title | SAMtools suite usage |
|---|
|
Program: samtools (Tools for alignments in the SAM format)
Version: 1.10 (using htslib 1.10)
Usage: samtools <command> [options]
Commands:
-- Indexing
dict create a sequence dictionary file
faidx index/extract FASTA
fqidx index/extract FASTQ
index index alignment
-- Editing
calmd recalculate MD/NM tags and '=' bases
fixmate fix mate information
reheader replace BAM header
targetcut cut fosmid regions (for fosmid pool only)
addreplacerg adds or replaces RG tags
markdup mark duplicates
-- File operations
collate shuffle and group alignments by name
cat concatenate BAMs
merge merge sorted alignments
mpileup multi-way pileup
sort sort alignment file
split splits a file by read group
quickcheck quickly check if SAM/BAM/CRAM file appears intact
fastq converts a BAM to a FASTQ
fasta converts a BAM to a FASTA
-- Statistics
bedcov read depth per BED region
coverage alignment depth and percent coverage
depth compute the depth
flagstat simple stats
idxstats BAM index stats
phase phase heterozygotes
stats generate stats (former bamcheck)
-- Viewing
flags explain BAM flags
tview text alignment viewer
view SAM<->BAM<->CRAM conversion
depad convert padded BAM to unpadded BAM |
In this exercise, we will explore five utilities provided by samtools: view, sort, index, flagstat, and idxstats. Each of these is executed in one line for a given SAM/BAM file. In the SAMtools/BEDtools sections tomorrow we will explore samtools in more in depth.
| Warning |
|---|
| title | Know your samtools version! |
|---|
|
There are two main "eras" of SAMtools development: - "original" samtools
- v 0.1.19 is the last stable version
- "modern" samtools
- v 1.0, 1.1, 1.2 – avoid these (very buggy!)
- v 1.3+ – finally stable!
Unfortunately, some functions with the same name in both version eras have different options and arguments! So be sure you know which version you're using. (The samtools version is usually reported at the top of its usage listing). TACC BioContainers also offers the original samtools version: samtools/ctr-0.1.19--3. |
samtools view
The samtools view utility provides a way of converting between SAM (text) and BAM (binary, compressed) format. It also provides many, many other functions which we will discuss lster. To get a preview, execute samtools view without any other arguments. You should see:
| Code Block |
|---|
|
Usage: samtools view [options] <in.bam>|<in.sam>|<in.cram> [region ...]
Options:
-b output BAM
-C output CRAM (requires -T)
-1 use fast BAM compression (implies -b)
-u uncompressed BAM output (implies -b)
-h include header in SAM output
-H print SAM header only (no alignments)
-c print only the count of matching records
-o FILE output file name [stdout]
-U FILE output reads not selected by filters to FILE [null]
-t FILE FILE listing reference names and lengths (see long help) [null]
-X include customized index file
-L FILE only include reads overlapping this BED FILE [null]
-r STR only include reads in read group STR [null]
-R FILE only include reads with read group listed in FILE [null]
-d STR:STR
only include reads with tag STR and associated value STR [null]
-D STR:FILE
only include reads with tag STR and associated values listed in
FILE [null]
-q INT only include reads with mapping quality >= INT [0]
-l STR only include reads in library STR [null]
-m INT only include reads with number of CIGAR operations consuming
query sequence >= INT [0]
-f INT only include reads with all of the FLAGs in INT present [0]
-F INT only include reads with none of the FLAGS in INT present [0]
-G INT only EXCLUDE reads with all of the FLAGs in INT present [0]
-s FLOAT subsample reads (given INT.FRAC option value, 0.FRAC is the
fraction of templates/read pairs to keep; INT part sets seed)
-M use the multi-region iterator (increases the speed, removes
duplicates and outputs the reads as they are ordered in the file)
-x STR read tag to strip (repeatable) [null]
-B collapse the backward CIGAR operation
-? print long help, including note about region specification
-S ignored (input format is auto-detected)
--no-PG do not add a PG line
--input-fmt-option OPT[=VAL]
Specify a single input file format option in the form
of OPTION or OPTION=VALUE
-O, --output-fmt FORMAT[,OPT[=VAL]]...
Specify output format (SAM, BAM, CRAM)
--output-fmt-option OPT[=VAL]
Specify a single output file format option in the form
of OPTION or OPTION=VALUE
-T, --reference FILE
Reference sequence FASTA FILE [null]
-@, --threads INT
Number of additional threads to use [0]
--write-index
Automatically index the output files [off]
--verbosity INT
Set level of verbosity |
That is a lot to process! For now, we just want to read in a SAM file and output a BAM file. The input format is auto-detected, so we don't need to specify it (although you do in v0.1.19). We just need to tell the tool to output the file in BAM format, and to include the header records.
| Expand |
|---|
|
| Code Block |
|---|
| language | bash |
|---|
| title | Get the alignment exercises files |
|---|
|
|
| Code Block |
|---|
| language | bash | mkdir -p $SCRATCH/core_ngs/ |
align2fastqyeast_bwa
cd $SCRATCH/core_ngs/ |
align2fastqcorral-repl/utexas/BioITeam/core_ngs_tools/alignment/*fastq.gz . |
|---|
Before executing the script you need to have one environment variable set. We'll do it at the command line here, but you could put it in your .bashrc file.
| Code Block |
|---|
|
export path_code=/work/01063/abattenh/code |
Now change into the directory and call the script with no arguments to see usage
catchup/yeast_bwa/yeast_pairedend.sam .
|
|
| Code Block |
|---|
| language | bash |
|---|
| title | Convert SAM to binary BAM |
|---|
|
| Code Block |
|---|
|
cd $SCRATCH/core_ngs/align2
$path_code/script/align/align_bwa_illumina.sh |
There are lots of bells and whistles in the arguments, but the most important are the first few:
- FASTQ file – full or relative path to the FASTQ file (just the R1 fastq if paired end). Can be compressed (.gz)
- output prefix – prefix for all the output files produced by the script. Should relate back to what the data is.
- assembly – genome assembly to use.
- there are pre-built indexes for some common eukaryotes (hg19, hg18, mm10, mm9, danRer7, sacCer3) that you can use
- or provide a full path for a bwa reference index you have built somewhere
- paired flag – 0 means single end (the default); 1 means paired end
- hard trim length – if you want the FASTQ hard trimmed down to a specific length, supply that number here
Now run the pipeline. By piping the output to tee <filename> we can see the script's progress at the terminal, and it also is written to <filename>.
| Code Block |
|---|
|
$path_code/script/align/align_bwa_illumina.sh ./fastq/Sample_Yeast_L005_R1.cat.fastq.gz yeast_chip sacCer3 1 2>&1 | tee aln.yeast_chip.log |
Output files
This alignment pipeline script performs the following steps:
- Hard trims FASTQ, if optionally specified (fastx_trimmer)
- Aligns the R1 FASTQ (bwa aln)
- Aligns the R2 FASTQ, if paired end alignment specified (bwa aln)
- Reports the alignments as SAM (bwa samse for single end, or bwa sampe for paired end)
- Converts SAM to BAM (samtools view)
- Sorts the BAM (samtools sort)
- Marks duplicates (Picard MarkDuplicates)
- Indexes the sorted, duplicate-marked BAM (samtools index)
- Gathers statistics (samtools idxstats, samtools flagstat, plus a custom statistics script of Anna's)
- Removes intermediate files
There are a number of output files, with the most important being those desribed below.
- aln.<prefix>.log – Log file of the entire alignment process.
- check the tail of this file to make sure the alignment was successful
- <prefix>.sort.dup.bam – Sorted, duplicate-marked alignment file.
- <prefix>.sort.dup.bam.bai – Index for the sorted, duplicate-marked alignment file
- <prefix>.samstats.txt – Summary alignment statistics from Anna's stats script
Verifying alignment success
The alignment log will have a "I ran successfully" message at the end if all went well, and if there was an error, the important information should also be at the end of the log file. So you can use tail to check the status of an alignment; for example:
| Code Block |
|---|
| language | bash |
|---|
| title | Checking the alignment log file |
|---|
|
tail aln.yeast_chip.log |
This will show something like:
| Code Block |
|---|
..samstats file 'yeast_chip.samstats.txt' exists Thu May 28 16:36:01 CDT 2015
..samstats file file 'yeast_chip.samstats.txt' size ok Thu May 28 16:36:01 CDT 2015
---------------------------------------------------------
Cleaning up files...
---------------------------------------------------------
ckRes 0 cleanup
---------------------------------------------------------
All bwa alignment tasks completed successfully!
Thu May 28 16:36:01 CDT 2015
--------------------------------------------------------- |
Notice that success message: "All bwa alignment tasks completed successfully!". It should only appear once in any successful alignment log.
When multiple alignment commands are run in parallel it is important to check them all, and you can use grep looking for part of the unique success message to do this.
For example, suppose I have run 6 alignments and have these 6 log files:
| Code Block |
|---|
aln.delswr1_htz1_tap1t0.log aln.delswr1_htz1_tap1t30.log aln.wt_htz1_tap1t15.log
aln.delswr1_htz1_tap1t15.log aln.wt_htz1_tap1t0.log aln.wt_htz1_tap1t30.log |
alignment/yeast_bwa
cat yeast_pairedend.sam | samtools view -b -o yeast_pairedend.bam |
- the -b option tells the tool to output BAM format
- the -o option specifies the name of the output BAM file that will be created
- we pipe the entire SAM file to samtools view so that the header records are included (required for SAM → BAM conversion)
- samtools view reads its input from standard input by default
How do you look at the BAM file contents now? That's simple. Just use samtools view without the -b option. Remember to pipe output to a pager!
| Code Block |
|---|
| language | bash |
|---|
| title | View BAM records |
|---|
|
samtools view yeast_pairedend.bam | more
|
Notice that this does not show us the header record we saw at the start of the SAM file.
Exercise: What samtools view option will include the header records in its output? Which option would show only the header records?
| Expand |
|---|
|
samtools view -h shows header records along with alignment records. samtools view -H shows header records only. |
samtools sort
Looking at some of the alignment record information (e.g. samtools view yeast_pairedend.bam | cut -f 1-4 | more), you will notice that read names appear in adjacent pairs (for the R1 and R2), in the same order they appeared in the original FASTQ file. Since that means the corresponding mappings are in no particular order, searching through the file very inefficient. samtools sort re-orders entries in the SAM file either by locus (contig name + coordinate position) or by read name.
If you execute samtools sort without any options, you see its help page:
| Code Block |
|---|
|
Usage: samtools sort [options...] [in.bam]
Options:
-l INT Set compression level, from 0 (uncompressed) to 9 (best)
-m INT Set maximum memory per thread; suffix K/M/G recognized [768M]
-n Sort by read name
-t TAG Sort by value of TAG. Uses position as secondary index (or read name if -n is set)
-o FILE Write final output to FILE rather than standard output
-T PREFIX Write temporary files to PREFIX.nnnn.bam
--no-PG do not add a PG line
--input-fmt-option OPT[=VAL]
Specify a single input file format option in the form
of OPTION or OPTION=VALUE
-O, --output-fmt FORMAT[,OPT[=VAL]]...
Specify output format (SAM, BAM, CRAM)
--output-fmt-option OPT[=VAL]
Specify a single output file format option in the form
of OPTION or OPTION=VALUE
--reference FILE
Reference sequence FASTA FILE [null]
-@, --threads INT
Number of additional threads to use [0]
--verbosity INT
Set level of verbosity |
In most cases you will be sorting a BAM file from name order to locus order. You can use either -o or redirection with > to control the output.
| Expand |
|---|
|
Copy aligned yeast BAM file | Code Block |
|---|
| mkdir -p $SCRATCH/core_ngs/alignment/yeast_bwa
cd $SCRATCH/core_ngs/alignment/yeast_bwa
cp $CORENGS/catchup/yeast_bwa/yeast_pairedend.bam . |
|
To sort the paired-end yeast BAM file by position, and get a BAM file named yeast_pairedend.sort.bam as output, execute the following I can check that all 6 completed with this command:
| Code Block |
|---|
| language | bash |
|---|
| title | Count the number of successful alignments |
|---|
|
grep 'completed successfully' aln.*.log | wc -l |
If this command returns 6, I'm done. But what if it doesn't? If you grep -v (lines that don't contain the pattern), you'll get every line in every log file except the success message line, which is not what you want at all.
...
|
cd $SCRATCH/core_ngs/alignment/yeast_bwa
samtools sort -O bam -T yeast_pairedend.tmp yeast_pairedend.bam > yeast_pairedend.sort.bam |
- The -O options says the Output format should be BAM
- The -T options gives a prefix for Temporary files produced during sorting
- sorting large BAMs will produce many temporary files during processing
- By default sort writes its output to standard output, so we use > to redirect to a file named yeast_pairedend.sort.bam
Exercise: Compare the file sizes of the yeast_pariedend .sam, .bam, and .sort.bam files and explain why they are different.
| Expand |
|---|
|
| grep |
| title | Count the number of successful alignments |
|---|
L 'completed successfully' aln.*.log |
The -L option tells grep to only print the filenames that don't contain the pattern. Perfect!
Checking alignment statistics
The <prefix>.samstats.txt statistics file produced by the alignment pipeline has a lot of good information in one place. If you use cat or more to view it you'll see this:
| Code Block |
|---|
-----------------------------------------------
Aligner: bwa
Total sequences: 1184360
Total mapped: 547664 (46.2 %)
Total unmapped: 636696 (53.8 %)
Primary: 547664 (100.0 %)
Secondary:
Duplicates: 324280 (59.2 %)
Fwd strand: 272898 (49.8 %)
Rev strand: 274766 (50.2 %)
Multi hit: 18688 (3.4 %)
Soft clip: 222451 (40.6 %)
All match: 319429 (58.3 %)
Indels: 6697 (1.2 %)
Spliced:
-----------------------------------------------
Total PE seqs: 1184360
PE seqs mapped: 547664 (46.2 %)
Num PE pairs: 592180
F5 1st end mapped: 300477 (50.7 %)
F3 2nd end mapped: 247187 (41.7 %)
PE pairs mapped: 241180 (40.7 %)
PE proper pairs: 236557 (39.9 %)
-----------------------------------------------
Insert size stats for: yeast_chip
Number of pairs: 236557 (proper)
Number of insert sizes: 212
Mean [-/+ 1 SD]: 215 [153 277] (sd 62)
Mode [Fivenum]: 223 [105 210 220 229 321]
----------------------------------------------- |
Since this was a paired end alignment there is paired-end specific information reported, including insert size statistics: mean/standard deviation, mode (most common insert size value) and fivenum (min, q1, median, q3 max insert sizes).
A quick way to check alignment stats if you have run multiple alignments is again to use grep. For example, for the 6 alignment files shown earlier, running this:
| Code Block |
|---|
| language | bash |
|---|
| title | Review multiple alignment rates |
|---|
|
grep 'Total map' *samstats.txt |
will produce output like this:
| Code Block |
|---|
delswr1_htz1_tap1t0.samstats.txt: Total mapped: 32761761 (86.8 %)
delswr1_htz1_tap1t15.samstats.txt: Total mapped: 33699464 (89.2 %)
delswr1_htz1_tap1t30.samstats.txt: Total mapped: 28441655 (87.6 %)
wt_htz1_tap1t0.samstats.txt: Total mapped: 28454847 (89.5 %)
wt_htz1_tap1t15.samstats.txt: Total mapped: 33245627 (90.9 %)
wt_htz1_tap1t30.samstats.txt: Total mapped: 32567026 (90.7 %) |
TACC batch system considerations
The great thing about pipeline scripts like this is that you can perform alignments on many datasets in parallel at TACC.
Anna's alignment pipeline scripts are written to take advantage of having multiple cores on TACC nodes, and are thus designed to run with at most two pipeline commands per TACC node.
| Tip |
|---|
| title | Always specify wayness 2 for these pipeline scripts |
|---|
|
These pipeline scripts should always be run with a wayness of 2 (-w 2) in the TACC batch system, meaning two commands per node. |
Assuming you have your alignment commands in a file called aln.cmds, here's how to create and submit a batch job for the commands.
| Code Block |
|---|
| language | bash |
|---|
| title | Submit BWA alignment pipeline job |
|---|
|
launcher_creator.py -n aln -j aln.cmds -t 12:00:00 -q normal -w 2
sbatch aln.slurm
showq -u |
Note the maximum run time specified here is 12 hours (-t 12:00:00). This is a reasonable value for a higher eukaryote with 20-40 M reads, and is way more than a yeast alignment would need (~ 4 hours). For very deeply sequenced eukaryotes (e.g. human genome re-sequencing with hundresd of millions of reads), you may want to specify the maximum job time of 48 hours.
Exercise: What would alignment commands look like if you were putting it in a batch system .cmds file?
| Expand |
|---|
|
Assuming you have $path_code set properly before submitting the job, the batch command would look like the command above, but you don't need the tee pipe. Instead, just redirect all output to a file. The example below shows how you would run alignments on two yeast samples in a batch file, adjusting the output prefix (yeast1, yeast2) and log file (aln.yeast1.log, aln.yeast2.log) accordingly. | Code Block |
|---|
| $path_code/script/align/align_bwa_illumina.sh ./fastq/Sample_Yeast_L005_R1.cat.fastq.gz yeast1 sacCer3 1 2>&1 > aln.yeast1.log
$path_code/script/align/align_bwa_illumina.sh ./fastq/Sample_ABCDE_L005_R1.cat.fastq.gz yeast2 sacCer3 1 2>&1 > aln.yeast2.log |
|
x
Exercise #2: Bowtie2 global alignment - Vibrio cholerae RNA-seq
While we have focused on aligning eukaryotic data, the same tools can be used to perform identical functions with prokaryotic data. The major differences are less about the underlying data and much more about the external/public databases established to store and distribute reference data. For example, the Illumina iGenome resource provides pre-processed and uniform reference data, designed to be out-of-the-box compatible with aligners like bowtie2 and bwa. However, the limited number of available species are heavily biased towards model eukaryotes. If we wanted to study a prokaryote, the reference data must be downloaded from a resource like GenBank, and processed/indexed similarly to the procedure for mirbase.
While the alignment procedure for prokaryotes is broadly analogous, the reference preparation process is somewhat different, and will involve use of a biologically-oriented scripting library called BioPerl. In this exercise, we will use some RNA-seq data from Vibrio cholerae, published last year on GEO here, and align it to a reference genome.
Overview of Vibrio cholerae alignment workflow with Bowtie2
Alignment of this prokaryotic data follows the workflow below. Here we will concentrate on steps 1 and 2.
- Prepare the vibCho reference index for bowtie2 from a GenBank record using BioPerl
- Align reads using bowtie2, producing a SAM file
- Convert the SAM file to a BAM file (samtools view)
- Sort the BAM file by genomic location (samtools sort)
- Index the BAM file (samtools index)
- Gather simple alignment statistics (samtools flagstat and samtools idxstat)
Obtaining the GenBank record(s)
V. cholerae has two chromosomes. We download each separately.
- Navigate to http://www.ncbi.nlm.nih.gov/nuccore/NC_012582
- click on the Send down arrow (top right of page)
- select Complete Record
- select Clipboard as Destination
- click Add to Clipboard
- Perform these steps in your Terminal window
- mkdir $WORK/tmp; cd $WORK/tmp
- type wget then paste the URL from the clipboard
- Repeat steps 1 and 2 fot the 2nd chromosome
- Combine the 2 files into one using cat
- cat NC_012582 NC_012583 > vibCho.gbk
Converting GenBank records into sequence (FASTA) and annotation (GFF) files
As noted earlier, many microbial genomes are available through repositories like GenBank that use specific file format conventions for storage and distribution of genome sequence and annotations. The GenBank file format is a text file that can be parsed to yield other files that are compatible with the pipelines we have been implementing.
Go ahead and look at some of the contents of a GenBank file with the following commands (execute these one at a time):
| Code Block |
|---|
|
cd $WORK/core_ngs/references
less vibCho.O395.gbk # use q to quit less
grep -A 5 ORIGIN vibCho.O395.gbk |
As the less command shows, the file begins with a description of the organism and some source information, and the contains annotations for each bacterial gene. The grep command shows that, indeed, there is sequence information here (flagged by the word ORIGIN) that could be exported into a FASTA file. There are a couple ways of extracting the information we want, namely the reference genome and the gene annotation information, but a convenient one (that is available through the module system at TACC) is BioPerl.
We load BioPerl like we have loaded other modules, with the caveat that we must load regular Perl before loading BioPerl:
| Code Block |
|---|
|
module load perl
module load bioperl |
These commands make several scripts directly available to you. The one we will use is called bp_seqconvert.pl, and it is a BioPerl script used to inter-convert file formats like FASTA, GBK, and others. This script produces two output files:
- a FASTA format file for indexing and alignment
- a GFF file (standing for General Feature Format) contains information about all genes (or, more generally, features) in the genome
- remember, annotations such as GFFs must always match the reference you are using
To see how to use the script, just execute:
| Code Block |
|---|
|
bp_seqconvert.pl |
Clearly, there are many file formats that we can use this script to convert. In our case, we are moving from genbank to fasta, so the commands we would execute to produce and view the FASTA files would look like this:
| Code Block |
|---|
|
cd $WORK/core_ngs/references
bp_seqconvert.pl --from genbank --to fasta < vibCho.O395.gbk > vibCho.O395.fa
mv vibCho.O395.fa fasta/
grep ">" fasta/vibCho.O395.fa
less fasta/vibCho.O395.fa |
Now we have a reference sequence file that we can use with the bowtie2 reference builder, and ultimately align sequence data against.
Recall from when we viewed the GenBank file that there are genome annotations available as well that we would like to extract into GFF format. However, the bp_seqconvert.pl script is designed to be used to convert sequence formats, not annotation formats. Fortunately, there is another script called bp_genbank2gff3.pl that can take a GenBank file and produce a GFF3 (the most recent format convention for GFF files) file. To run it and see the output, run these commands:
| Code Block |
|---|
|
bp_genbank2gff3.pl --format Genbank vibCho.O395.gbk
mv vibCho.O395.gbk.gff vibCho.O395.gff
less vibCho.O395.gff |
After the header lines, each feature in the genome is represented by a line that gives chromosome, start, stop, strand, and other information. Features are things like "mRNA," "CDS," and "EXON." As you would expect in a prokaryotic genome it is frequently the case that the gene, mRNA, CDS, and exon annotations are identical, meaning they share coordinate information. You could parse these files further using commands like grep and awk to extract, say, all exons from the full file or to remove the header lines that begin with #.
Introducing bowtie2
Go ahead and load the bowtie2 module so we can examine some help pages and options. To do that, you must first load the perl module, and then the a specific version of bowtie2.
| Code Block |
|---|
|
module load perl
module load bowtie/2.2.0
|
Now that it's loaded, check out the options. There are a lot of them! In fact for the full range of options and their meaning, Google "Bowtie2 manual" and bring up that page. The Table of Contents is several pages long! Ouch!
This is the key to using bowtie2 - it allows you to control almost everything about its behavior, but that also makes it is much more challenging to use than bwa – and it's easier to screw things up too!
Building the bowtie2 vibCho index
Before the alignment, of course, we've got to build a mirbase index using bowtie2-build (go ahead and check out its options). Unlike for the aligner itself, we only need to worry about a few things here:
- reference_in file is just the FASTA file containing mirbase v20 sequences
- bt2_index_base is the prefix of where we want the files to go
To build the reference index for alignment, we actually only need the FASTA file, since annotations are often not necessary for alignment. (This is not always true - extensively spliced transcriptomes requires splice junction annotations to align RNA-seq data properly, but for now we will only use the FASTA file.)
| Code Block |
|---|
|
mkdir -p $WORK/core_ngs/references/bt2/vibCho
mv $WORK/core_ngs/references/vibCho.O395.fa $WORK/core_ngs/references/fasta
cd $WORK/core_ngs/references/bt2/vibCho
ln -s -f ../../fasta/vibCho.O395.fa
ls -la |
Now build the index using bowtie2-build:
| Code Block |
|---|
| language | bash |
|---|
| title | Prepare Bowtie2 index files |
|---|
|
bowtie2-build vibCho.O395.fa vibCho.O395 |
This should also go pretty fast. You can see the resulting files using ls like before.
Performing the bowtie2 alignment
Now we will go back to our scratch area to do the alignment, and set up symbolic links to the index in the work area to simplify the alignment command:
| Code Block |
|---|
|
cd $SCRATCH/core_ngs/alignment
ln -s -f $WORK/core_ngs/references/bt2/vibCho vibCho |
Note that here the data is from standard mRNA sequencing, meaning that the DNA fragments are typically longer than the reads. There is likely to be very little contamination that would require using a local rather than global alignment, or many other pre-processing steps (e.g. adapter trimming). Thus, we will run bowtie2 with default parameters, omitting options other than the input, output, and reference index.
As you can tell from looking at the bowtie2 help message, the general syntax looks like this:
| Code Block |
|---|
bowtie2 [options]* -x <bt2-idx> {-1 <m1> -2 <m2> | -U <r>} [-S <sam>] |
So our command would look like this:
| Code Block |
|---|
|
bowtie2 -x vibCho/vibCho.O395 -U fastq/cholera_rnaseq.fastq.gz -S cholera_rnaseq.sam |
| Expand |
|---|
|
Parameters are: - -x vibCho/vibCho.O395.fa – prefix path of index files
- -U fastq/cholera_rnaseq.fastq.gz – FASTQ file for single-end (Unpaired) alignment
- -S cholera_rnaseq.sam – tells bowtie2 to report alignments in SAM format to the specified file
|
Create a commands file called bt2_vibCho.cmds with this task definition then generate and submit a batch job for it (time 1 hour, development queue).
| Expand |
|---|
|
Use nano to create the bt2_vibCho.cmds file. Then: | Code Block |
|---|
| language | bash |
|---|
| title | Local bowti2 alignment of miRNA data |
|---|
| launcher_creator.py -n bt2_vibCho -j bt2_vibCho.cmds -t 01:00:00 -A UT-2015-05-18
sbatch bt2_vibCho.slurm; showq -u |
|
...
| Expand |
|---|
|
The yeast_pairedend.sam text file is the largest at ~348 MB. The name-ordered binary yeast_pairedend.bam text file only about 1/3 that size, ~110 MB. They contain exactly the same records, in the same order, but conversion from text to binary results in a much smaller file. The coordinate-ordered binary yeast_pairedend.sort.bam file is even slightly smaller, ~91 MB. This is because BAM files are actually customized gzip-format files. The customization allows blocks of data (e.g. all alignment records for a contig) to be represented in an even more compact form. You can read more about this in section 4 of the SAM format specification. |
samtools index
Many tools (like IGV, the Integrative Genomics Viewer) only need to use portions of a BAM file at a given point in time. For example, if you are viewing alignments that are within a particular gene, alignment records on other chromosomes do not need to be loaded. In order to speed up access, BAM files are indexed, producing BAI files which allow fast random access. This is especially important when you have many alignment records.
The utility samtools index creates an index that has the same name as the input BAM file, with suffix .bai appended. Here's the samtools index usage:
| Code Block |
|---|
| title | samtools index usage |
|---|
|
Usage: samtools index [-bc] [-m INT] <in.bam> [out.index]
Options:
-b Generate BAI-format index for BAM files [default]
-c Generate CSI-format index for BAM files
-m INT Set minimum interval size for CSI indices to 2^INT [14]
-@ INT Sets the number of threads [none] |
The syntax here is way, way easier. We want a BAI-format index which is the default. (CSI-format is used with extremely long contigs, which don't apply here - the most common use case is for polyploid plant genomes).
So all we have to provide is the sorted BAM:
| Code Block |
|---|
| language | bash |
|---|
| title | Index a sorted bam |
|---|
|
samtools index yeast_pairedend.sort.bam |
This will produce a file named yeast_pairedend.bam.bai.
Most of the time when an index is required, it will be automatically located as long as it is in the same directory as its BAM file and shares the same name up until the .bai extension.
Exercise: Compare the sizes of the sorted BAM file and its BAI index.
| Expand |
|---|
|
| Code Block |
|---|
| ls -lh yeast_pairedend.sort.bam* |
|
| Expand |
|---|
|
While the yeast_pairedend.sort.bam text file is ~91 MB, its index (yeast_pairedend.sort.bai) is only 20 KB. |
samtools flagstat
Since the BAM file contains records for both mapped and unmapped reads, just counting records doesn't provide information about the mapping rate of our alignment. The samtools flagstat tool provides a simple analysis of mapping rate based on the the SAM flag fields.
Here's how to run samtools flagstat and both see the output in the terminal and save it in a file – the samtools flagstat standard output is piped to tee, which both writes it to the specified file and sends it to its standard output:
| Code Block |
|---|
| language | bash |
|---|
| title | Run samtools flagstat using tee |
|---|
|
samtools flagstat yeast_pairedend.sort.bam | tee yeast_pariedend.flagstat.txt |
You should see something like this:
| Code Block |
|---|
| title | samtools flagstat output |
|---|
|
1184360 + 0 in total (QC-passed reads + QC-failed reads)
0 + 0 secondary
0 + 0 supplementary
0 + 0 duplicates
547664 + 0 mapped (46.24% : N/A)
1184360 + 0 paired in sequencing
592180 + 0 read1
592180 + 0 read2
473114 + 0 properly paired (39.95% : N/A)
482360 + 0 with itself and mate mapped
65304 + 0 singletons (5.51% : N/A)
534 + 0 with mate mapped to a different chr
227 + 0 with mate mapped to a different chr (mapQ>=5) |
Ignore the "+ 0" addition to each line - that is a carry-over convention for counting QA-failed reads that is no longer relevant.
The most important statistic is the mapping rate (here 46%) but this readout also allows you to verify that some common expectations (e.g. that about the same number of R1 and R2 reads aligned, and that most mapped reads are proper pairs) are met.
Exercise: What proportion of mapped reads were properly paired?
| Expand |
|---|
|
Divide the number of properly paired reads by the number of mapped reads: | Code Block |
|---|
| awk 'BEGIN{ print 473114 / 547664 }'
# or
echo $(( 473114 * 100 / 547664 ))
|
|
| Expand |
|---|
|
About 86% of mapped read were properly paired. This is actually a bit on the low side for ChIP-seq alignments which typically over 90%. |
samtools idxstats
More information about the alignment is provided by the samtools idxstats report, which shows how many reads aligned to each contig in your reference. Note that samtools idxstats must be run on a sorted, indexed BAM file.
| Code Block |
|---|
| language | bash |
|---|
| title | Use samtools idxstats to summarize mapped reads by contig |
|---|
|
samtools idxstats yeast_pairedend.sort.bam | tee yeast_pairedend.idxstats.txt |
| Code Block |
|---|
| language | bash |
|---|
| title | samtools idxstats output |
|---|
|
chrI 230218 8820 1640
chrII 813184 36616 4026
chrIII 316620 13973 1530
chrIV 1531933 72675 8039
chrV 576874 27466 2806
chrVI 270161 10866 1222
chrVII 1090940 50893 5786
chrVIII 562643 24672 3273
chrIX 439888 16246 1739
chrX 745751 31748 3611
chrXI 666816 28017 2776
chrXII 1078177 54783 10124
chrXIII 924431 40921 4556
chrXIV 784333 33070 3703
chrXV 1091291 48714 5150
chrXVI 948066 44916 5032
chrM 85779 3268 291
* 0 0 571392 |
The output has four tab-delimited columns:
- contig name
- contig length
- number of mapped reads
- number of unmapped reads
The reason that the "unmapped reads" field for named chromosomes is not zero is that the aligner may initially assign a potential mapping (contig name and start coordinate) to a read, but then mark it later as unampped if it does meet various quality thresholds.
| Tip |
|---|
If you're mapping to a non-genomic reference such as miRBase miRNAs or another set of genes (a transcriptome), samtools idxstats gives you a quick look at quantitative alignment results. |


