This page should serve as a reference for the many "things Linux" we use in this course.
Getting around in the shell
Important keyboard shortcuts
Type as little and as accurately as possible by using keyboard shortcuts!
Tab key completion
The Tab key is your best friend! Hit the Tab key once or twice - it's almost always magic! Hitting Tab invokes shell completion, instructing the shell to try to guess what you're doing and finish the typing for you. On most modern Linux shells, Tab completion will:
- single Tab – complete file or directory names up to any ambiguous part
- if nothing shows up, there is no unambiguous match
- Tab twice – display all possible completions
- you then decide where to go next
- work for shell commands too (like ls or cp)
Arrow keys
- Use Up arrow to retrieve any of the last 500 commands you've typed, going backwards through your history.
- You can then edit them and hit Enter (even in the middle of the command) and the shell will use that command.
- The Down arrow "scrolls" forward from where you are in the command history.
- Right arrow and Left arrow move the cursor forward or backward on the current command line.
Command line editing
- Use Ctrl-a (holding down the "control" key and "a") to jump the cursor to the beginning of the line.
- Use Ctrl-e to jump the cursor to the end of the line.
- Arrow keys are also modified by Ctrl
- e.g. Ctrl-right-arrow will skip by word forward; Ctrl-left-arrow backward.
- Use Backspace to remove text before the cursor; Delete to remove text after the cursor.
Wildcards and special file names
The shell has shorthand to refer to groups of files by allowing wildcards in file names.
* (asterisk) is the most common filename wildcard. It matches "any length of any characters".
This technique is sometimes called filename globbing, and the pattern a glob.
Other useful ones are brackets ( [ ] ) to allow for any character in the list of characters between the brackets. And you can use a hyphen ( - ) to specify a range of characters
For example:
- ls *.bam – lists all files in the current directory that end in .bam
- ls [A-Z]*.bam – does the same, but only if the first character of the file is a capital letter
- ls [ABcd]*.bam – lists all .bam files whose 1st letter is A, B, c or d.
Three special file names:
.(single period) means "this directory"...(two periods) means "directory above current." So ls .. means "list contents of the parent directory."- ~ (tilde) means "my home directory".
Avoid spaces in filenames
While it is possible to create file and directory names that have embedded spaces, that creates problems when manipulating them.
To avoid headaches, it is best not to create file/directory names with embedded spaces.
Standard streams
Every command and Linux program has three "built-in" streams: standard input, standard output and standard error.
It is easy to not notice the difference between standard output and standard error when you're in an interactive Terminal session – because both outputs are sent to the Terminal. But they are separate streams, with different meanings. When running batch programs and scripts you will want to manipulate standard output and standard error from programs appropriately.
redirecting output
- To take the standard output of a program and save it to a file, you use the > operator
- a single > overwrites any existing target; a double >> appends to it
- since standard output is stream #1, this is the same as 1>
- To redirect the standard error of a program you must specify its stream number using 2>
- To redirect standard output and standard error to the same place, use the syntax 2>&1
To see the difference between standard output and standard error try these commands:
# redirect a long listing of your $HOME directory to a file ls -la $HOME > cmd.out # look at the contents -- you'll see just files cat cmd.out # this command gives an error because the target does not exist ls -la bad_directory # redirect any errors from ls to a file ls -la bad_directory 2> cmd.out # look at the contents -- you'll see an error message cat cmd.out # now redirect both error and output streams to the same place ls -la bad_directory $HOME > cmd.out # look at the contents -- you'll see both an error message and files cat cmd.out
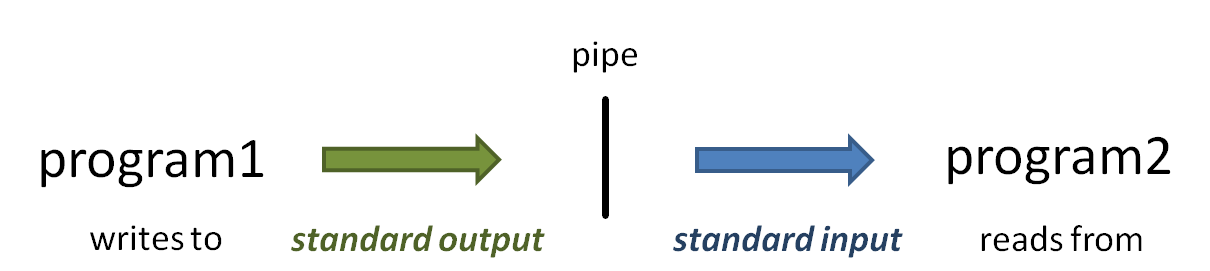
piping
The power of the Linux command line is due in no small part to the power of piping. The pipe symbol ( | ) connects one program's standard output to the next program's standard input.
A simple example is piping uncompressed data "on the fly" to a pager like more (or less):
# zcat is like cat, except that it understands the gz compressed format, # and uncompresses the data before writing it to standard output. # So, like cat, you need to be sure to pipe the output to a pager if # the file is large. zcat big.fq.gz | more # Another way to do the same thing is to use gunzip and provide the -c option, # which says to write decompressed data to the stdout (-c for "console") gunzip -c big.fq.gz | more
piping a histogram
But the real power of piping comes when you stitch together a string of commands with pipes – it's incredibly flexible, and fun once you get the hang of it.
For example, here's a simple way to make a histogram of mapping quality values from a subset of BAM file records.
# create a histogram of mapping quality scores for the 1st 1000 mapped bam records samtools view -F 0x4 small.bam | head -1000 | cut -f 5 | sort -n | uniq -c
- samtools view converts the binary small.bam file to text and writes alignment record lines one at a time to standard output.
- -F 0x4 option says to filter out any records where the 0x4 flag bit is 0 (not set)
- since the 0x4 flag bit is set (1) for unmapped records, this says to only report records where the query sequence did map to the reference
- | head -1000
- the pipe connects the standard output of samtools view to the standard input of head
- the -1000 option says to only write the first 1000 lines of input to standard output
- | cut -f 5
- the pipe connects the standard output of head to the standard input of cut
- the -f 5 option says to only write the 5th field of each input line to standard output (input fields are tab-delimited by default)
- the 5th field of an alignment record is an integer representing the alignment mapping quality
- the resulting output will have one integer per line (and 1000 lines)
- | sort -n
- the pipe connects the standard output of cut to the standard input of sort
- the -n option says to sort input lines according to numeric sort order
- the resulting output will be 1000 numeric values, one per line, sorted from lowest to highest
- | uniq -c
- the pipe connects the standard output of sort to the standard input of uniq
- the -c option option says to just count groups of lines with the same value (that's why they must be sorted) and report the total for each group
- the resulting output will be one line for each group that uniq sees
- each line will have the text for the group (here the unique mapping quality values) and a count of lines in each group
Environment variables
Environment variables are just like variables in a programming language (in fact bash is a complete programming language), they are "pointers" that reference data assigned to them. In bash, you assign an environment variable as shown below:
export varname="Some value, here it's a string"
Careful – do not put spaces around the equals sign when assigning environment variable values.
Also, always use double quotes if your value contains (or might contain) spaces.
You set environment variables using the bare name (varname above).
You then refer to or evaluate an environment variables using a dollar sign ( $ ) before the name:
echo $varname
The export keyword when you're setting ensures that any sub-processes that are invoked will inherit this value. Without the export only the current shell process will have that variable set.
Use the env command to see all the environment variables you currently have set.
Quoting in the shell
What different quote marks mean in the shell and when to use can be quite confusing.
There are three types of quoting in the shell:
- single quoting (e.g. 'some text') – this serves two purposes
- it groups together all text inside the quotes into a single argument that is passed to the command
- it tells the shell not to "look inside" the quotes to perform any evaluations
- any environment variables in the text – or anything that looks like an environment variable – are not evaluated
- no pathname globbing (e.g. *) is performed
- double quoting (e.g. "some text") – also serves two purposes
- it groups together all text inside the quotes into a single argument that is passed to the command
- it inhibits pathname globbing, but allows environment variable evaluation
- backtick quoting (e.g. `date`)
- evaluates the expression inside the backticks
- the resulting standard output of the expression replaces the backticked text
Using Commands
Command options
Sitting at the computer, you should have some idea what you need to do. There's probably a command to do it. If you have some idea what it starts with, you can type a few characters and hit Tab twice to get some help. If you have no idea, you Google it or ask someone else.
Once you know a basic command, you'll soon want it to do a bit more - like seeing the sizes of files in addition to their names.
Most built-in commands in Linux use a common syntax to ask more of a command. They usually add a dash ( - ) followed by a code letter that names the added function. These "command line switches" are called options.
Options are, well, optional – you only add them when you need them. The part of the command line after the options, like filenames, are called arguments. Arguments can also be optional, but you can tell them from options because they don't start with a dash.
# long listing option (-l) ls -l # long listing (-l), all files (-a) and human readable file sizes (-h) options. $HOME is an argument (directory name) ls -l -a -h $HOME # sort by modification time (-t) displaying a long listing (-l) that includes the date and time ls -lt
Almost all built-in Linux commands, and especially NGS tools, use options heavily.
Like dialects in a language, there are at least three basic schemes commands/programs accept options in:
Single-letter short options, which start with a single dash ( - ) and can often be combined, like:
Examples of different short optionshead -20 # show 1st 20 lines ls -lhtS (equivalent to ls -l -h -t -S)
Long options use the convention that double dashes ( -- ) precede the multi-character option name, and they can never be combined. Strictly speaking, long options should be separated from their values by the equals sign ( = ) according to the POSIX standard (see https://en.wikipedia.org/wiki/POSIX). But most programs let you use a space as separator also. Here's an example using the mira genome assembler:
Example of long optionsmira --project=ct --job=denovo,genome,accurate,454 -SK:not=8
Word options, illustrated in the GATK command line to call SNPs below.
Word options combine aspects of short and long options – they usually start with a single dash ( - ), but can be multiple letters and are never combined.
Sometimes the option (e.g. java's -Xms initial memory heap size option), and its value (512m which means 512 megabytes) may be smashed together.
Other times a multi-letter switch and its value are separated by a space (e.g. -glm BOTH).
java -Xms512m -Xmx4g -jar /work/projects/BioITeam/common/opt/GenomeAnalysisTK.jar -glm BOTH -R $reference -T UnifiedGenotyper -I $outprefix.realigned.recal.bam --dbsnp $dbsnp -o $outprefix.snps.vcf -metrics snps.metrics -stand_call_conf 50.0 -stand_emit_conf 10.0 -dcov 1000 -A DepthOfCoverage -A AlleleBalance
Getting help
So you've noticed that options can be complicated – not to mention program arguments. Some options have values and others don't. Some are short, others long. How do you figure out what kinds of functions a command (or NGS tool) offers? You need help!
--help option
Many (but not all) built-in shell commands will give you some help if you provide the long --help option. This can often be many pages, so you'll probably want to pipe the output to a pager like more. This is most useful to remind yourself what the name of that dang option was, assuming you know something about it.
-h or -? options
The -h and -? options are similar to --help. If --help doesn't work, try -h. or -?. Again, output can be lengthy and best used if you already have an idea what the program does.
just type the program name
Many 3rd party tools will provide extensive usage information if you just type the program name then hit Enter.
For example:
module load bwa bwa
Produces something like this:
Program: bwa (alignment via Burrows-Wheeler transformation)
Version: 0.7.12-r1039
Contact: Heng Li <lh3@sanger.ac.uk>
Usage: bwa <command> [options]
Command: index index sequences in the FASTA format
mem BWA-MEM algorithm
fastmap identify super-maximal exact matches
pemerge merge overlapping paired ends (EXPERIMENTAL)
aln gapped/ungapped alignment
samse generate alignment (single ended)
sampe generate alignment (paired ended)
bwasw BWA-SW for long queries
shm manage indices in shared memory
fa2pac convert FASTA to PAC format
pac2bwt generate BWT from PAC
pac2bwtgen alternative algorithm for generating BWT
bwtupdate update .bwt to the new format
bwt2sa generate SA from BWT and Occ
Note: To use BWA, you need to first index the genome with `bwa index'.
There are three alignment algorithms in BWA: `mem', `bwasw', and
`aln/samse/sampe'. If you are not sure which to use, try `bwa mem'
first. Please `man ./bwa.1' for the manual.
Notice that bwa, like many NGS programs, is written as a set of sub-commands. This top-level help displays the sub-commands available. You then type bwa <command> to see help for the sub-command:
bwa index
Displays something like this:
Usage: bwa index [options] <in.fasta>
Options: -a STR BWT construction algorithm: bwtsw or is [auto]
-p STR prefix of the index [same as fasta name]
-b INT block size for the bwtsw algorithm (effective with -a bwtsw) [10000000]
-6 index files named as <in.fasta>.64.* instead of <in.fasta>.*
Warning: `-a bwtsw' does not work for short genomes, while `-a is' and
If you don't already know much about a command (or NGS tool), just Google it! Try something like "bwa manual" or "rsync man page". Many tools have websites that combine tool overviews with detailed option help. Even for built-in Linux commands, you're likely to get hits of a tutorial style, which are more useful when you're getting started.
And it's so much easier to read things in a nice web browser!
man pages
Linux has had built-in help files since the mid-1500's, way before Macs or PCs thought of such things. They're called man pages (short for manual).
For example, man intro will give you an introduction to all user commands.
man pages will detail all options available – in excruciating detail (unless there's no man page ), so the manual system has its own built-in pager. The pager is sort of like less, but not quite the same (why make it easy?). We recommend man pages only for advanced users.
Basic linux commands you need to know
Here's a copy of the cheat sheet we passed out.
And here's a set of commands you should know, by category (under construction).
Command line arguments can be replaced by standard input
Most built-in Linux commands that obtain data from command line arguments (such as file names) can also accept the data piped in on their standard input.
File system navigation
- ls - list the contents of the speicified directory
- -l says produce a long listing (including file permissions, sizes, owner and group
- -a says show all files, even normally-hidden dot files whose names start with a period ( . )
- -h says to show file sizes in human readable form (e.g. 12M instead of 12201749)
- cd <whereto> - change the current working directory to <whereto>. Some special <wheretos>:
- .. (period, period) means "up one level"
- ~ (tilde) means "my home directory"
- file <file> tells you what kind of file <file> is
- df shows you the top level directory structure of the system you're working on, along with how much disk space is available
- -h says to show sizes in human readable form (e.g. 12G instead of 12318201749)
- pwd - display the present working directory
- -P says to display the full absolute path
Create, rename, link to, delete files
- touch <file> – create an empty file, or update the modification timestamp on an existing file
- mkdir -p <dirname> – create directory <dirname>.
- -p says to create any needed subdirectories also
- mv <file1> <file2> – renames <file1> to <file2>
- mv <file1> <file2> ... <fileN> <dir>/ – moves files <file1> <file2> ... <fileN> into directory <dir>
- ln -s <path> creates a symbolic (-s) link to <path> in the current directory
- default link name corresponds to the last name component in <path>
- always change into (cd) the directory where you want the link before executing ln -s
- a symbolic link can be deleted without affecting the linked-to file
- rm <file> deletes a file. This is permanent - not a "trash can" deletion.
- rm -rf <dirname> deletes an entire directory – be careful!
Displaying file contents
- cat outputs all the contents of its input (one or more files and/or standard input) or the specified file
- CAUTION – only use on small files!
- zcat <file.gz> like cat, but understands the gzip (.gz) format, and decompresses the data before writing it to standard output
- CAUTION – only use on small files!
- Another CAUTION – does not understand .zip or .bz2 compression formats
- more and less "pagers"
- both display their (possibly very long) input one Terminal "page" at a time
- in more:
- use spacebar to advance a page
- use q or Ctrl-c to exit more
- in less:
- q – quit
- Ctrl-f or space – page forward
- Ctrl-b – page backward
- /<pattern> – search for <pattern> in forward direction
- n – next match
- N – previous match
- ?<pattern> – search for <pattern> in backward direction
- n – previous match going back
- N – next match going forward
- use less -N to display line numbers
- can be used directly on .gz files
- head and tail
- show you the top or bottom 10 lines (by default) of their input
- head -20 show the top 20 lines
- tail -2 shows the last 2 lines
- tail -n +100 shows lines starting at line 100
- tail -f shows the last lines of a file, then follows the output as more lines are written (Ctrl-c to quit)
- gunzip -c <file.gz> | more (or less) – like zcat, un-compresses lines of <file.gz> and outputs them to standard output
- <file.gz> is not altered on disk
- always pipe the output to a pager!
Copying files and directories
- cp <source> <destination> copies the file <source> to the location and/or file name <destination>.
- using . (period) as the destination means "here, with the same name"
- -p option says to preserve file times
- cp -r <dirname> <destination> will recursively copy the directory dirname and all its contents to the directory destination.
- scp <user>@<host>:<source_path> <destination_path>
- Works just like cp but copies source from the a directory on the remote host machine to the local file destination
- -p (preserve file times) and -r (recursive) options work the same as cp
- A nice scp syntax resource is located here.
- wget <url> fetches a file from a valid URL (e.g. http, https, ftp).
- -O <file> specifies the name for the local file (defaults to the last component of the URL)
- rsync -arvP <source_directory>/ <target_directory>/
rsync -ptlrvP <source_directory>/ <target_directory>/- Recursively copies <source_directory> contents to <target_directory>, but only if <source_directory> files are newer or don't yet exist in <target_directory>
Remote path syntax (<user>@<host>:<absolute_or_home-relative_path>) can be used for either source or target (but not both).
- Always include a trailing slash ( / ) after the source and target directory names
- -p means preserve file permissions
- -t means preserve file times
- -l means copy symbolic links as links (vs -L which means dereference the link and copy the file it refers to)
- -a means "archive" mode (equivalent to -ptl and some other options)
- -r means recursively copy sub-directories
- -v means verbose
- -P means show transfer Progress (useful when large files are being transferred)
Miscellaneous commands
- wc -l reports the number of lines (-l) in its input
- history lists your command history to the terminal
- redirect to a file to save a history of the commands executed in a shell session
- pipe to grep to search for a particular command
- which <pgm> searches all $PATH directories to find <pgm> and reports its full pathname
Copying files from TACC to your laptop
Assume you want to copy the TACC file $SCRATCH/core_ngs/fastq_prep/small_fastqc.html back to your laptop.
First, figure out what the appropriate absolute path (a.k.a. full pathname) is on TACC.
cd $SCRATCH/core_ngs/fastq/prep pwd -P
This will return something like /scratch/01063/abattenh/core_ngs/fastq_prep.
For folks with Mac or Linux laptops, just open a terminal window, cd to the directory where you want the files, and type something like following, substituting your user name and absolute path:
scp abattenh@ls5.tacc.utexas.edu:/scratch/01063/abattenh/core_ngs/fastq_prep/small_fastqc.html .
For Windows users, pscp.exe, a remote file copy program should have been installed with Putty. To use it, first open a Command window (Start menu, search for Cmd). Then in the Command window, see if it is on your Windows %PATH% by just typing the executable name:
pscp.exe
If this shows usage information, you're good to go. Execute something like following, substituting your user name and absolute path:
cd c:\Scratch pscp.exe abattenh@ls5.tacc.utexas.edu:/scratch/01063/abattenh/core_ngs/fastq_prep/small_fastqc.html .
If pscp.exe is not on your %PATH%, you may need to locate the program. Try this:
cd "c:\Program Files"\putty dir
If you see the program pscp.exe, you're good. You just have to use its full path. For example:
cd c:\Scratch "c:\Program Files"\putty\pscp.exe abattenh@ls5.tacc.utexas.edu:/scratch/01063/abattenh/core_ngs/fastq_prep/small_fastqc.html
Advanced commands
- cut -f <field_number(s)> extracts one or more fields (-f) from each line of its input
- -d <delim> to change the field delimiter (tab by default)
- sort sorts its input using an efficient algorithm
- by default sorts each line lexically, but one or more fields to sort can be specified with one or more -k <field_number> options
- options to sort numerically (-n), or numbers-inside-text (version sort -V)
- uniq -c counts groupings of its input (which must be sorted) and reports the text and count for each group
- use cut | sort | uniq -c for a quick-and-dirty histogram (see piping a histogram)
- grep -P '<pattern>' searches for <pattern> in its input and outputs only lines containing it
- always enclose <pattern> in single quotes to inhibit shell evaluation!
- -P says use Perl patterns, which are much more powerful than standard grep patterns
- <pattern> can contain special match "meta-characters" such as:
- ^ – matches beginning of line
- $ – matches end of line
- . – (period) matches any single character
- \s – matches any whitespace (\S any non-whitespace)
- \d – matches digits 0-9
- \w – matches any word character: A-Z, a-z, 0-9 and _ (underscore)
- \t matches Tab; \r matches Carriage return; \n matches Linefeed
- [xyz123] – matches any single character (including special characters) among those listed between the brackets [ ] x
- this is called a character class.
- use [^xyz123] to match any single character not listed in the class
- (Xyz|Abc) – matches either Xyz or Abc or any text or expressions inside parentheses separated by | characters
- note that parentheses ( ) may also be used to capture matched sub-expressions for later use
- * – modifier; place after an expression to match 0 or more occurrences
- + – modifier, place after an expression to match 1 or more occurrences
- Regular expression modules are available in nearly every programming language (Perl, Python, Java, PHP, awk, even R)
- each "flavor" is slightly different
- even bash has multiple regex commands: grep, egrep, fgrep.
- There are many good online regular expression tutorials, but be sure to pick one tailored to the language you will use.
- here's a good general one: https://www.regular-expressions.info/
- awk '<script>' a powerful scripting language that is easily invoked from the command line
- <script> is applied to each line of input (generally piped in)
- always enclose <script> in single quotes to inhibit shell evaluation
- General structure of an awk script:
- {BEGIN <expressions>} – use to initialize variables before any script body lines are executed
- e.g. {BEGIN FS="\t"; OFS="\t"; sum=0} says
- use tab (\t) as the input (FS) and output (OFS) field separator (default is a space), and
- initialize the variable sum to 0.
- e.g. {BEGIN FS="\t"; OFS="\t"; sum=0} says
- {<body expressions>} – expressions to apply to each line of input
- use $1, $2, etc. to pick out specific input fields
- e.g. {print $3,$4} outputs fields 3 and 4 of the input
- {END <expressions>} – executed after all input is complete (e.g. print a sum)
- {BEGIN <expressions>} – use to initialize variables before any script body lines are executed
- Here is an excellent awk tutorial, very detailed and in-depth
- take a look once you feel comfortable with the example scripts we've gone over in class.
- <script> is applied to each line of input (generally piped in)
calculate average insert size
Here is an example awk script that works in conjunction with samtools view to calculate the average insert size for properly paired reads in a BAM file produced by a paired-end alignment:
samtools view -F 0x4 -f 0x2 yeast_pairedend.sort.bam | awk \
'BEGIN{ FS="\t"; sum=0; nrec=0; }
{ if ($9 > 0) {sum += $9; nrec++;} }
END{ print sum/nrec; }'
- samtools view converts each alignment record in yeast_pairedend.sort.bam to text
- the -F 0x4 filter says to output records only for mapped sequences (ones assigned a contig and position)
- BAM files often contain records for both mapped and unmapped reads
- -F filters out records where the specified bit(s) are not set (i.e., they are 0)
- so technically we're asking for "not unmapped" reads since bit 0x4 = 1 means unmapped
- the -f 0x2 filter says to output only reads that are flagged as properly paired by the aligner
- these are reads where both R1 and R2 reads mapped within a "reasonable" genomic distance
- -f filters out records where the specified bit(s) are set (i.e., they are 1)
- alignment records that pass both filters are written to standard output
- the -F 0x4 filter says to output records only for mapped sequences (ones assigned a contig and position)
- | awk \
- the pipe | connects the standard output of samtools view to the standard input of awk
- the backslash character \ is simply a line continuation marker
- allows us to split a long command line like this into multiple text lines for clarity
- be careful not to add any spaces or tabs after the backslash
- 'BEGIN{ ... }'
- these 3 lines of text, enclosed in single quotes, are the awk script
- we don't have to use line continuation characters within the script because the newline characters within the quotes are part of the script
- the BEGIN block between the curly brackets { } is executed once before the script processes any input data
- it says to use tab ("\t") as the input (FS) field separator (default is a space), and initialize the variables sum and nrec to 0.
- it says to use tab ("\t") as the input (FS) field separator (default is a space), and initialize the variables sum and nrec to 0.
- { if ($9 > 0) {sum += $9; nrec++} }
- this is the body of the awk script, which is executed for each line of input
- $9 represents the 9th tab-delimited field of the input
- the 9th field of an alignment record is the insert size, according to the SAM format spec
- we only execute the main part of the body when the 9th field is positive: if ($9 > 0)
- since each proper pair will have one alignment record with a positive insert size and one with a negative insert size, this check keeps us from double-counting insert sizes for pairs
- when the 9th field is positive, we add its value to sum (sum += $9) and add one to our record count (nrec++)
- END{ print sum/nrec; }
- the END block between the curly brackets { } is executed once after the script has processed all input data
- this prints the average insert size (sum/nrec) to standard output
- these 3 lines of text, enclosed in single quotes, are the awk script
process multiple files with a for loop
The general structure of a for loop in bash are shown below. Different portions of the structure can be separated on different lines (like <something> and <something else> below) or put on one line separated with a semicolon ( ; ) like before the do keyword below.
for <variable name> in <expression>; do <something> <something else> done
One common use of for loops is to process multiple files, where the set of files to process is obtained by pathname wildcarding. For example, the code below
for fname in *.gz; do echo "$fname has $((`zcat $fname | wc -l` / 4)) sequences" done
Here fname is the name given the variable that is assigned a different filename each time through the loop. The set of such files is generated by the filename wildcard matching *.gz. The actual file is then referenced as $fname inside the loop.
Editing files
There are several options for editing files at TACC. These fall into three categories:
- Linux command-line text editors installed at TACC (nano, vi, emacs). These run in your Terminal window.
- nano is extremely simple and is the best choice as a first local text editor
- vi and emacs are extremely powerful but also quite complex
- Text editors or IDEs that run on your local computer but have an SFTP (secure FTP) interface that lets you connect to a remote computer
- E.g., Notepad++ or Komodo Edit
- Once you connect to the remote host, you can navigate its directory structure and edit files.
- When you open a file, its contents are brought over the network into the text editor's edit window, then saved back when you save the file.
- Software that will allow you to mount your home directory on TACC as if it were a normal disk
- then, you can use any text editor or IDE on your local computer to open/edit/save files (although it will be slower than local file editing)
- e.g. ExpanDrive for Windows or Mac (costs $$, but has a free trial), TextWrangler for Mac.
Knowing the basics of at least one Linux text editor is useful for creating small files like TACC commands files. We'll use nano for this in class.
For editing larger files, you may find options #2 or #3 more useful.
nano
nano is a very simple editor available on most Linux systems. If you are able to ssh into a remote system, you can use nano there.
To invoke nano to edit a new or existing file just type:
nano <filename>
You'll see a short menu of operations at the bottom of the terminal window. The most important are:
- Ctrl-o - write out the file
- Ctrl-x - exit nano
You can just type in text, and navigate around using arrow keys. A couple of other navigation shortcuts:
- Ctrl-a - go to start of line
- Ctrl-e - go to end of line
Be careful with long lines – sometimes nano will split long lines into more than one line, which can cause problems in commands files
Line ending nightmares
The dirty little secret of the computer world is that the three main "families" of computers – Macs, Windows and Linux/Unix – use different, mutually incompatible line endings.
- Linux/Unix uses linefeed ( \n )
- Windows uses carriage return followed by linefeed ( \r\n )
- some Mac programs use carriage return only ( \r )
And guess what? Most Linux programs don't work with files that have Windows or Mac line endings, and what's worse they give you bizarre error messages that don't give you a clue what's going on.
So whatever non-Linux text editor you use, be sure to adjust its "line endings" setting – and it better have one somewhere!
Komodo Edit for Mac and Windows
Komodo Edit is a free, full-featured text editor with syntax coloring for many programming languages and a remote file editing interface. It has versions for both Macintosh and Windows. Download the appropriate install image here.
Once installed, start Komodo Edit and follow these steps to configure it:
- Configure the default line separator for Unix
- On the Edit menu select Preferences
- Select the New Files Category
- For Specify the end-of-line (EOL) indicator for newly created files select UNIX (\n)
- Select OK
- Configure a connection to TACC
- On the Edit menu select Preferences
- Select the Servers Category
- For Server type select SFTP
- Give this profile the Name of ls5
- For Hostname enter ls5.tacc.utexas.edu
- Enter your TACC user ID for Username
- Leave Port and Default path blank
- Select OK
When you want to open an existing file at Lonestar5, do the following:
- Select the File menu -> Open -> Remote File
- Select your ls5 profile from the top Server drop-down menu
- Once you log in, it should show you all the files and directories in your ls5 $HOME directory
- Navigate to the file you want and open it
- Often you will use your ~/work or ~/scratch directory links to help you here
To create and save a new file, do the following:
- From the Komodo Edit Start Page, select New File
- Select the file type (Text is good for commands files)
- Edit the contents
- Select the File menu -> Save As Other -> Remote File
- Select your ls5 profile from the Server drop-down menu
- Once you log in, it should show you all the files and directories in your ls5 $HOME directory
- Navigate to where you want the put the file and save it
- Often you will use your ~/work or ~/scratch directory links to help you here
Rather than having to navigate around TACC's complex file system tree, it helps to use the symbolic links to those areas that we created in your home directory.
Notepad++ for Windows
Notepad++ is an open source, full-featured text editor for Windows PCs (not Macs). It has syntax coloring for many programming languages (python, perl, shell), and a remote file editing interface.
If you're on a Windows PC download the installer here.
Once it has been installed, start Notepad++ and follow these steps to configure it:
- Configure the default line separator for Unix
- In the Settings menu, select Preferences
- In the Preferences dialog, select the New Document/Default Directory tab.
- Select Unix in the Format section
- Close
- Configure a connection to TACC
- In the Plugins menu, select NppFTP, then select Focus NppFTP Window. The top bar of the NppFTP panel should become blue.
- Click the Settings icon (looks like a gear), then select Profile Settings
- In the Profile settings dialog click Add new
- Call the new profile ls5
- Fill in Hostname (ls5.tacc.utexas.edu) and your TACC user ID
- Connection type must be SFTP
- Close
To open the connection, click the blue (Dis)connect icon then select your ls5 connection. It should prompt for your password. Once you've authenticated, a directory tree ending in your home directory will be visible in the NppFTP window. You can click the the (Dis)connect icon again to Disconnect when you're done.
Rather than having to navigate around TACC's complex file system tree, it helps to use the symbolic links to those areas that we created in your home directory (~/work or ~/scratch).