Overview
As we have seen, the SAMTools suite allows you to manipulate the SAM/BAM files produced by most aligners. There are many sub-commands in this suite, but the most common and useful use cases are:
- Convert text-format SAM files into binary BAM files (samtools view) and vice versa
- Sort BAM files by reference coordinates (samtools sort)
- Index BAM files that have been sorted (samtools index)
- Filter alignment records based on BAM flags, mapping quality or location
Since BAM files are binary, they can't be viewed directly using standard Unix file viewers such as more, less and head. We have seen how samtools view can be used to binary-format BAM files into text format for viewing. But samtools view also has options that let you do powerul filtering of the output. We focus on this filtering capability in this set of exercises.
Setup
Set up a directory for these exercises and copy an indexed BAM file there. This is a renamed version of the yeast_pairedend.sort.bam file from our Alignment workflow exercises.
mkdir -p $SCRATCH/core_ngs/samtools cd $SCRATCH/core_ngs/samtools cp $CORENGS/catchup/for_samtools/* .
References
Handy links
- The SAM format specification
- especially section 1.4 - alignment section fields
- Manual for SAMTools 1.3.1
- especially the 1st section on samtools view.
SAM header fields
The 11 SAM alignment record required fields (Tab-delimited).
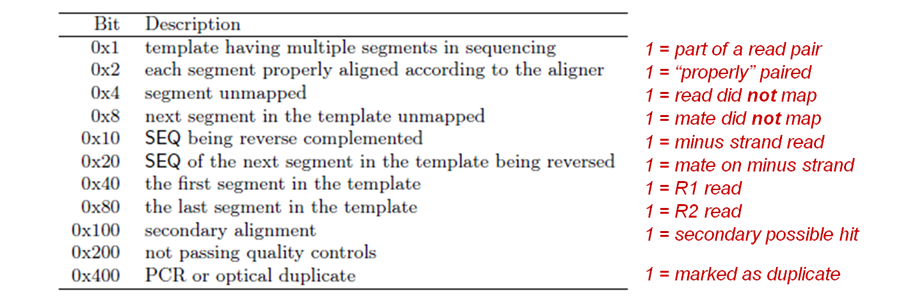
SAM flags field
Meaning of each bit (flag) in the SAM alignment records flags field (column 2). The most commonly querried flags are denoted in red.
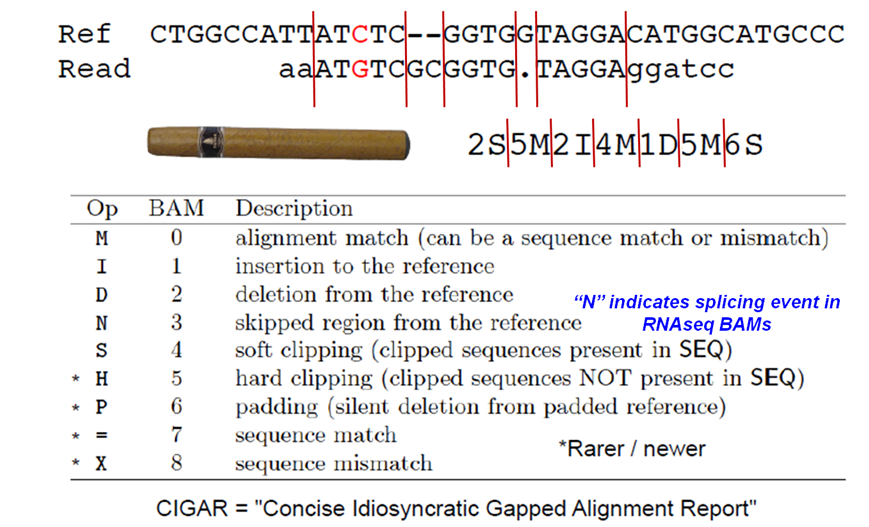
SAM CIGAR string
Format of the CIGAR string in column 6 of SAM alignment records.
Exercises
Analyzing the CIGAR string
Filtering for only mapped reads (samtools view -F 0x4)
The code block below details some basic samtools functionality:
samtools view -o outfile_view.bam infile.bam # use the -c option to just count alignment records samtools sort infile.bam outfile.sorted.bam samtools index aln.sorted.bam
First, logon to stampede and copy the file yeast_pairedend.bam to your $SCRATCH directory:
ssh user@login8.stampede.tacc.utexas.edu cd $SCRATCH/core_ngs mkdir samtools cd samtools cp $SCRATCH/core_ngs/alignment/yeast_pairedend.bam .
Samtools flags and mapping rate: calculating the proportion of mapped reads in an aligned bam file
We have a sorted, indexed BAM file. Now we can use other samtools functionality to filter this file and count mapped vs unmapped reads in a given region. samtools allows you to sort based on certain flags that are specified on page 4 on the sam format specification. We'll focus on a couple, below.
Here are three of the most useful flags to sort on. We'll be using the unmapped flag.
# SAM specifications common flag usage 0x04 = unmapped 0x02 = part of a properly aligned pair 0x400 = optical duplicate # look at samtools rmdup if you need to remove these sequences
Exercise 2: count unmapped reads vs total reads on chromosome III for the yeast_pairedend_sort.bam file you created above. What proportion of the reads are mapped?
Filtering bam files based on mapped status and mapping quality using samtools view
Mapping qualities are a measure of how likely a given sequence alignment to a location is correct. The lowest score is a mapping quality of zero, or mq0 for short. The reads map to multiple places on the genome, and we can't be sure of where the reads originated. To improve the quality of our data, we can remove these low quality reads from our sorted and indexed file.
Exercise 3: Remove unmapped and low quality reads from your bam file.