...
- -q N – only report alignment records with mapping quality of at least N (>= N).
- -f 0xXX – only report alignment records where the specified flags XX are all set (are all 1)
- you can provide the flags in decimal, or as here as hexidecimal
- -F 0xXX – only report alignment records where the specified flags XX are all cleared (are all 0)
Setup
Login to login5.ls5stampede2.tacc.utexas.edu, set up a directory for these exercises, and copy an indexed BAM file there. This is a renamed version of the yeast_pairedend.sort.bam file from our Alignment workflow exercises and start an idev session.
| Code Block |
|---|
| language | bash |
|---|
| title | Setup for samtools exercises | Start an idev session |
|---|
|
idevmkdir -p $SCRATCH/core_ngs/samtools
cd $SCRATCH/core_ngs/samtools
cp $CORENGS/catchup/for_samtools/* . |
References
Handy links
- The SAM format specification
- especially section 1.4 - alignment section fields
- Manual for SAMTools
- especially the 1st section on samtools view.
The 11 SAM alignment record required fields (Tab-delimited).
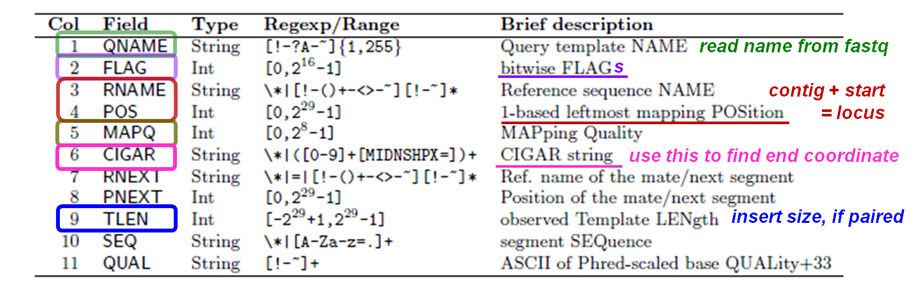 Image Removed
Image Removed
SAM flags field
development -m 180 -A UT-2015-05-18 -N 1 -n 68 --reservation=BIO_DATA_week_1 |
Next set up a directory for these exercises, and copy an indexed BAM file there. This is a renamed version of the yeast_pairedend.sort.bam file from our Alignment workflow exercises.
| Code Block |
|---|
| language | bash |
|---|
| title | Setup for samtools exercises |
|---|
|
mkdir -p $SCRATCH/core_ngs/samtools
cd $SCRATCH/core_ngs/samtools
cp $CORENGS/catchup/for_samtools/* . |
References
Handy links
- The SAM format specification
- especially section 1.4 - alignment section fields
- Manual for SAMTools
- especially the 1st section on samtools view.
The 11 SAM alignment record required fields (Tab-delimited).
 Image Added
Image Added
SAM flags field
Meaning of each bit (Meaning of each bit (flag) in the SAM alignment records flags field (column 2). The most commonly flags are denoted in red.
...
We'll do this first exercise one step at a time to get comfortable with piping commands. Start by just looking at the first few alignment records of the BAM file:
| Code Block |
|---|
|
cd $SCRATCH/core_ngs/alignment/samtools
module load samtools biocontainers # takes a while
module load samtools
cd $SCRATCH/core_ngs/alignment/samtools
samtools view yeast_pe.sort.bam | head |
...
| Code Block |
|---|
|
samtools view yeast_pe.sort.bam | cut -f 6 | head 20 |
Next, make sure we're only looking at alignment records that represent mapped reads. The -F 0x4 option says to filter records where the 0x4 flag (read unmapped) is 0, resulting it only mapped reads being output.
| Code Block |
|---|
|
samtools view -F 0x4 yeast_pe.sort.bam | cut -f 6 | head 20 |
Now we use grep with the pattern '[ID]' to select lines (which are now just CIGAR strings) that have Insert or Deletion operators. The brackets ( [ ] ) denote a character class pattern, which matches any of the characters inside the brackets. Be sure to ask for Perl regular expressions (-P) to be sure that this character class syntax is recognized.
| Code Block |
|---|
|
samtools view -F 0x4 yeast_pe.sort.bam | cut -f 6 | grep -P '[ID]' | head |
Ok, this looks good. We're ready to run this against all the alignment records, and count the results:
...
| Code Block |
|---|
| language | bash |
|---|
| title | One-liner for calculating BAM alignment rate |
|---|
|
echo "`samtools view -F 0x4 -c yeast_pe.sort.bam` `samtools view -F 0x4 yeast_pe.sort.bam | cut -f 6 | grep -P '[ID]' | wc -l`" | \
awk '{ print 100 * $2/$1,"%" }' 100 * $2/$1,"%" }' |
| Tip |
|---|
awk also has a printf function, which can take the standard formatting commands (see https://en.wikipedia.org/wiki/Printf_format_string#Type_field). | Code Block |
|---|
| awk 'BEGIN{ printf("%.2f %%\n", 100*6697/547664) } |
Notes: - The printf arguments are enclosed in parentheses since it is a true function.
- The 1st argument is the format string, enclosed in double quotes.
- The %.2f format specifier says to output a floating point number with 2 digits after the decimal place.
- The %% format specifier is used to output a single, literal percent sign.
- Unlike the standard print statement, the printf function does not automaticlly append a newline to the output, so \n is added to the format string here.
- Remaining arguments to printf are the values to be substituted for the format specifiers.
- Here just our percentage computation
|
Filtering by location range
...
| Expand |
|---|
|
| Code Block |
|---|
| mkdir -p $SCRATCH/core_ngs/samtools
cd $SCRATCH/core_ngs/samtools
cp $CORENGS/catchup/for_samtools/* .
module load biocontainers
module load samtools |
|
| Code Block |
|---|
|
cd $SCRATCH/core_ngs/alignment/samtools
# count the number of reads mapped to chromosome 2 (chrII)
samtools view -c -F 0x4 yeast_pe.sort.bam chrII
# count the number of reads mapped to chromosomes 1 or M (chrI, chrM)
samtools view -c -F 0x4 yeast_pe.sort.bam chrI chrM
# count the number of reads mapped to chromosomes 1 that overlap coordinates 1000-2000
samtools view -c -F 0x4 yeast_pe.sort.bam chrI:1000-2000
# since there are only 20 reads in the chrI:1000-2000 region, examine them individually
samtools view -F 0x4 yeast_pe.sort.bam chrI:1000-2000
# look at a subset of field for chrI:1000-2000 reads
# 2=flags, 3=contig, 4=start, 5=mapping quality, 6=CIGAR, 9=insert size
samtools view -F 0x4 yeast_pe.sort.bam chrI:1000-2000 | cut -f 2-6,9 |
...
| Code Block |
|---|
|
# look at a subset of field for chrI:1000-2000 reads
# 2=flags, 3=contig, 4=start, 5=mapping quality, 6=CIGAR, 9=insert size
samtools view -F 0x4 yeast_pe.sort.bam chrI:1000-2000 | cut -f 2-6,9 | \
awk 'BEGIN{FS=OFS="\t"}{$1 = sprintf("0x%x", $1); print}' |
Notes:
- Usually, if If a command line is continued on more than one line, the line continuation character ( \ ) is used.here we don't need a line continuation character because the linefeed after the first single quote is part of the script.
- There is a sprintf (string print formatted) function in awk, just as there is in many modern programming languages.
- We use sprintf to re-format the 1st input field (here the cut flags) in hex
- using the format directive (%x)
- also adding a literal "0x" prefix to denote the numeric base is hexadecimal
- The results of sprintf are stored back into the 1st field ($1), replacing the original value.
- awk's print statement is then used with no other arguments to print all its input fields, including the changed $1.
...
| Expand |
|---|
| title | Answer 1 (building on the last command) |
|---|
|
Here's one way to do it, building on our last command line: | Code Block |
|---|
| samtools view -F 0x4 yeast_pe.sort.bam chrI:1000-2000 | awk '
BEGIN{FS=OFS="\t"}
{$2 = sprintf("0x%x", $2); print $2}' | grep -c '3$' |
Note that here we don't need the line continuation character because the newline after the first single quote is part of the script command string. |
| Expand |
|---|
| title | Hint 2 (using a SAM flag) |
|---|
|
| Use the 0x2 flag (1 = properly paired) |
...
- bwa aln (global alignment) and bowtie2 with default parameters (both --local and --global) report at most one location for a read that maps
- this will be the location with the best mapping quality and alignment
- if a given read maps equally well to multiple locations, these aligners pick one location at random
- bwa aln will always report a 0 mapping quality for these multi-hit reads
- bowtie2 will report a low mapping quality (< 10), based on the complexity (information content) of the sequence
- bwa mem (local alignment) can always report more than one location for a mapped read
- its definition of a secondary alignment is different (and a bit non-standard)
- if one part of a read maps to one location and another part maps somewhere else (e.g. because of RNA splicing), the longer alignment is marked as primary and the shorter as secondary.
- there is no way to disable reporting of secondary alignments with bwa mem.
- but they can be filtered from the sorted BAM with -F 0x100 (secondary alignment flag = 0).
- bowtie2 can be configured to report more than one location for a mapped read
- the -k N option says to report up to N alignments for a read
- most transcriptome-aware RNA-seq aligners by default report more than one location for a mapped read
- e.g. tophat2, hisat2, star, tophat2.
- when reads are quantified (counted with respect to genes), multiply-mapped reads can be counted fractionally
- e.g. if a read maps to 5 genes, it can be counted as 1/5 for each of the genes
...
| Expand |
|---|
|
| Code Block |
|---|
| mkdir -p $SCRATCH/core_ngs/samtools
cd $SCRATCH/core_ngs/samtools
cp $CORENGS/catchup/for_samtools/* .
module load biocontainers
module load samtools |
|
Exercise: Use samtools view with -F, -f and -q options to create a BAM containing only mapped, properly paired, high-quality (mapQ 20+) reads. Call the output file yeast_pe.sort.filt.bam.
...
