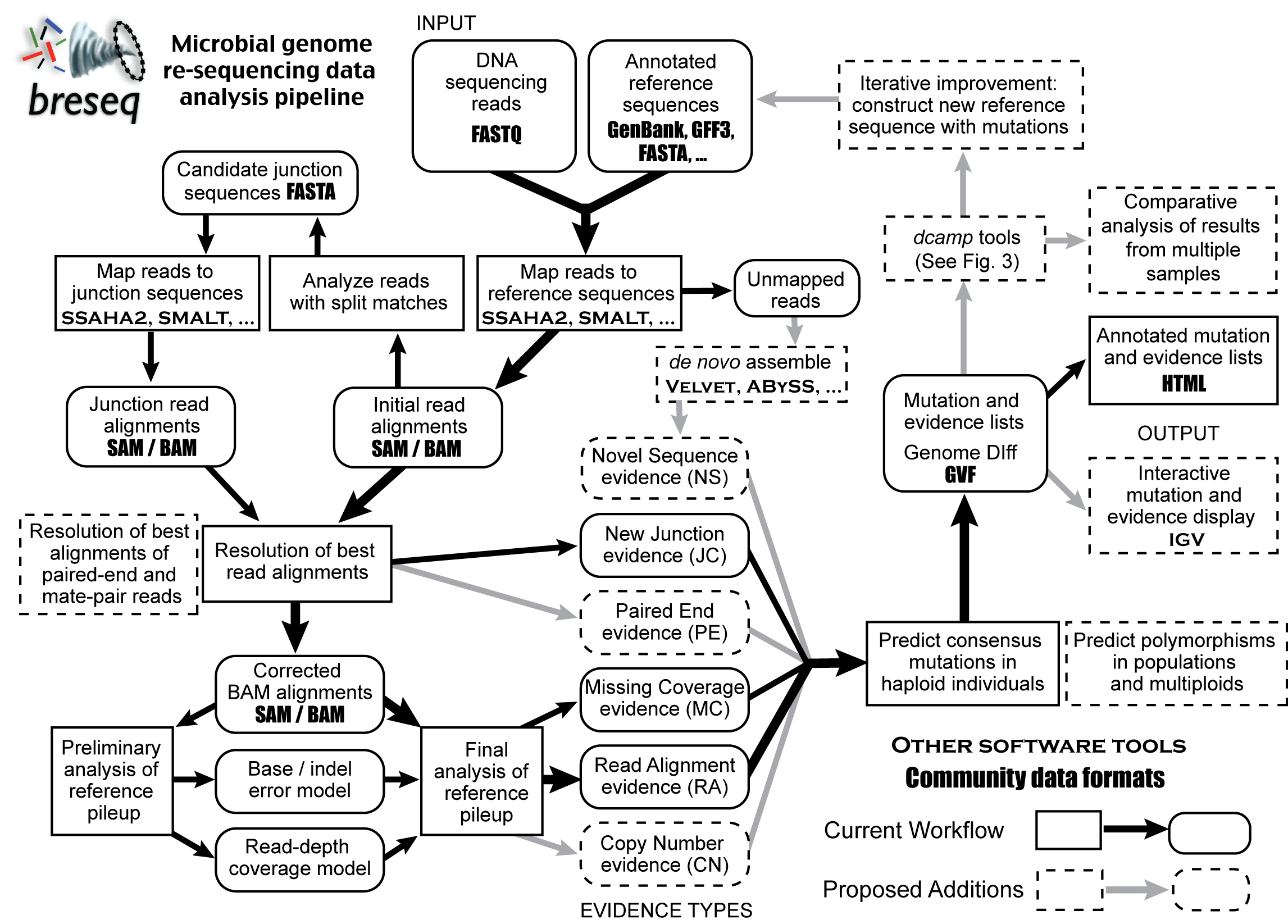
Introduction
breseq is a tool developed by the Barrick lab intended for analyzing genome re-sequencing data for bacteria. It is primarily used to analyze laboratory evolution experiments with microbes. In these experiments, there is usually a high-quality reference genome for the ancestral strain, and one is interested in exhaustively finding all of the mutations that occurred during the evolution experiment. Then one might want to construct a phylogenetic tree of individuals samples from a single population or determine whether the same gene is mutated in many independent evolution experiments in an environment.
Input data / expectations:
- Haploid reference genome
- Relatively small (<20 Mb) reference genome
- Input FASTQ reads can be from any sequencing technology
- Average genomic coverage > 30-fold
- Less than ~1,000 mutations expected
- Detects SNVs and SVs from single-end reads (does not use paired-end distance information)
- Produces annotated HTML output
You can learn a great deal more about breseq by reading the Online Documentation.
Here is a rough outline of the workflow in breseq with proposed additions.
Table of Contents
Optional: Install breseq
We have already installed breseq in $BI/bin, so you should be ready to go! Skip this section.
Download breseq from Google code
See if you can install breseq and get it running from the installation instructions.
Info for installing breseq on TACC
You do not need to install a compiler (GCC/ICC), bowtie2, or R on TACC as they are available via the module system.
module load R module load GCC module load bowtie/2.1.0
You could add these commands to your $HOME/.profile_user if you wanted them available by default.
Example 1: Bacteriophage lambda data set
First, we'll run breseq on a small data set to be sure that it is installed correctly, and to get a taste for what the output looks like. This sample is a mixed population of bacteriophage lambda that was co-evolved in lab with its E. coli hosts.
Data
The data files for this example are in the path:
$BI/ngs_course/lambda_mixed_pop/data
Copy this directory to your $SCRATCH space. Name it something other than data. And cd into it.
File Name | Description | Sample |
|---|---|---|
| Single-end Illumina 36-bp reads | Evolved lambda bacteriophage mixed population genome sequencing |
| Reference Genome | Bacteriophage lambda |
Running breseq
Because this data set is relatively small (roughly 100x coverage of a 48,000 bp genome), a breseq run will take < 5 minutes. Submit this command to the TACC development queue.
breseq -j 12 -r lambda.gbk lambda_mixed_population.fastq > log.txt
A bunch of progress messages will stream by during the breseq run. They detail several steps in a pipeline that combines the steps of mapping (using SSAHA2), variant calling, annotating mutations, etc. You can examine them by peeking in the log.txt file as your job runs using tail -f. The -f option means to "follow" the file and keep giving you output from it as it gets bigger. You will need to wait for your job to start running before you can tail -f log.txt.
Looking at breseq predictions
breseq will produce a lot of directories beginning 01_sequence_conversion, 02_reference_alignment, ... Each of these contains intermediate files that can be deleted when the run completes, or explored if you are interested in the inner guts of what is going on.
breseq will also produce two directories called: data and output.
First, copy the output directory back to your desktop computer.
Inside of the output directory is a file called index.html. Open this in a web browser on your desktop and click around to take a look at the mutation predictions and summary information.
Example 2: E. coli data sets
Now we'll try running breseq on some Escherichia coli genomes from an evolution experiment. These files are larger. You don't want to run them in interactive mode. We'll submit them to the TACC queue all at once.
Data
The data files for this example are in the path:
$BI/ngs_course/ecoli_clones/data
File Name | Description | Sample |
|---|---|---|
| Paired-end Illumina 36-bp reads | 0K generation evolved E. coli strain |
| Paired-end Illumina 36-bp reads | 2K generation evolved E. coli strain |
| Paired-end Illumina 36-bp reads | 5K generation evolved E. coli strain |
| Paired-end Illumina 36-bp reads | 10K generation evolved E. coli strain |
| Paired-end Illumina 36-bp reads | 15K generation evolved E. coli strain |
| Paired-end Illumina 36-bp reads | 20K generation evolved E. coli strain |
| Paired-end Illumina 36-bp reads | 40K generation evolved E. coli strain |
| Reference Genome | E. coli B str. REL606 |
Running breseq on TACC
breseq may take an hour or two to run on these sequences, so you should submit to the serial queue instead of the development queue on TACC and should give a run time of 4 hours as a conservative estimate.
Since we have multiple data sets, this example will also give us an opportunity to run several commands as part of a single job on TACC, and use multiple cores on a single processor.
You'll want each command to look something like this:
login1$ breseq -r NC_012967.1.gbk -o output_00K SRR030252_1.fastq SRR030252_2.fastq
Notice the additional -o option which specifies that all of those output directories should be put in the specified directory, instead of the current directory. If we don't include this, then we will end up writing the output from all of the runs on top of one other. The program will undoubtedly get confused, possibly crash, and generally it will be a mess.
Hint: It is often a good idea to try running a command that you are about to submit to the TACC queue yourself, just to be sure you have all the options and paths correct. Otherwise you will have to wait until it starts running on TACC in order to find out that it it failed immediately, which can be frustrating. Try running the command above. You can use control-c to cancel the command after you're sure it started ok.
Once you have your commands file ready, then you need to create your launcher.sge script.
Examining breseq results
As before, copy the data back to your computer and examine the HTML output in a web browser.
Exercise: Can you figure out how to archive all of the output directories and copy only those files (and not all of the very large intermediate files) back to your machine? - without deleting any files?
Click around in the results.
Optional: breseq utility commands
breseq includes a few utility commands that can be used on any BAM/FASTA set of files to draw an HTML read pileup or a plot of the coverage across a region.
It's easiest to run these commands from inside the main output directory (e.g., output_20K) of a breseq run. They use information in the data directory.
breseq bam2aln NC_012967:237462-237462 breseq bam2cov NC_012967:2300000-2320000
Additionally, the files in the data directory can be loaded in IGV if you copy them back to your desktop.
Optional Exercise: Running breseq in mixed population mode
The phage lambda data set you examined is actually a mixed population of many different phage lambda genotypes descended from a clonal ancestor. You ran breseq in a mode where it predicted consensus mutations in what it thinks is one uniform haploid genome. Actually, some individuals in the population have certain mutations and others do not, so you might have noticed when you looked at some of the alignments that there was a mixture of bases at a position.
We will talk more about analyzing mixed population data to predict rare variants in a later lesson. However, if you're curious you can now experimental with running breseq in a mode where it estimates the frequencies of different mutations in the population. This process is most accurate for single nucleotide variants. Mutations at intermediate frequencies are not (yet) predicted for all classes of mutations like large structural variants.
login1$ breseq --polymorphism-prediction --polymorphism-no-indels -r lambda.gbk lambda_mixed_population.fastq
The option --polymorphism-prediction turns on these mixed population predictions. The option --polymorphism-no-indels turns off predictions of small insertions and deletions (which don't work as well for reasons too complicated to explain here). You're welcome to also try it without this option.
Copy the resulting output directory back to your computer and examine the HTML output in a web browser. Compare it to the output from before.