...
Since BAM files are binary, they can't be viewed directly using standard Unix file viewers such as more, less and head. We have seen how samtools view can be used to binary-format BAM files into text format for viewing. But samtools view also has options that let you do powerul filtering of the output. We focus on this filtering capability in this set of exercises.
Setup
Set Login to login5.ls5.tacc.utexas.edu and set up a directory for these exercises and copy an indexed BAM file there. This is a renamed version of the yeast_pairedend.sort.bam file from our Alignment workflow exercises.
| Code Block | ||||
|---|---|---|---|---|
| ||||
mkdir -p $SCRATCH/core_ngs/samtools cd $SCRATCH/core_ngs/samtools cp $CORENGS/catchup/for_samtools/* . |
References
Handy links
- The SAM format specification
- especially section 1.4 - alignment section fields
- Manual for SAMTools 1.3.1
- especially the 1st section on samtools view.
...
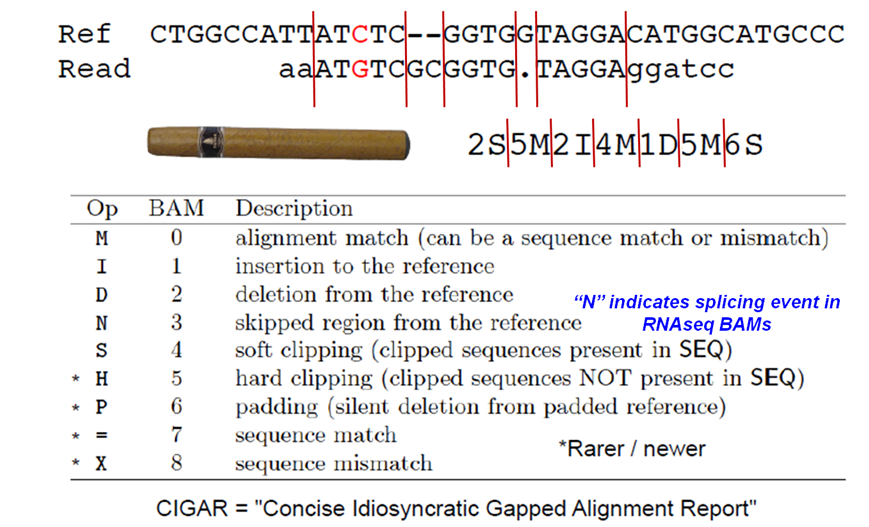
Format of the CIGAR string in column 6 of SAM alignment records.
Exercises
Analyzing the CIGAR string for indels
Suppose we want to know how many alignments included insertions or deletions (indels) versus the reference. Looking at the CIGAR field definition table above, we see that insertions are denoted with the character I and deletions with the character D. We'll use this information, along with samtools view, cut and grep, to count the number of indels.
We'll do this first exercise one step at a time to get comfortable with piping commands. Start by just looking at the first few alignment records of the BAM file:
| Code Block | ||
|---|---|---|
| ||
cd $SCRATCH/core_ngs/alignment/samtools
module load samtools
samtools view yeast_pe.sort.bam | head |
With all the line wrapping, it looks pretty ugly. Still, you can pick out the CIGAR string in colum 6. Let's select just that column with cut:
| Code Block | ||
|---|---|---|
| ||
samtools view yeast_pe.sort.bam | cut -f 6 | head |
Next, make sure we're only looking at alignment records that represent mapped reads. The -F 0x4 option says to filter records where the 0x4 flag (read unmapped) is 0, resulting it only mapped reads being output.
| Code Block | ||
|---|---|---|
| ||
samtools view -F 0x4 yeast_pe.sort.bam | cut -f 6 | head |
Now we use grep with the pattern '[ID]' to select lines (which are now just CIGAR strings) that have Insert or Deletion operators. The brackets ( [ ] ) denote a character class pattern, which matches any of the characters inside the brackets. Be sure to ask for Perl regular expressions (-P) so that this character class syntax is recognized.
| Code Block | ||
|---|---|---|
| ||
samtools view -F 0x4 yeast_pe.sort.bam | cut -f 6 | grep -P '[ID]' | head |
Ok, this looks good. We're ready to run this against all the alignment records, and count the results:
| Code Block | ||
|---|---|---|
| ||
samtools view -F 0x4 yeast_pe.sort.bam | cut -f 6 | grep -P '[ID]' | wc -l |
There are 6697 such records.
What is that in terms of the rate of indels? For that we need to count the total number of mapped reads. Here we can just use the -c (count only) option to samtools view.
| Code Block | ||
|---|---|---|
| ||
samtools view -F 0x4 -c yeast_pe.sort.bam |
x
x
x
Filtering for only mapped reads (samtools view -F 0x4)
...